Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces
- PMID: 23183971
- PMCID: PMC3568538
- DOI: 10.1128/AEM.03157-12
Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces
Abstract
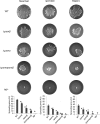
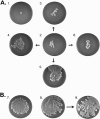
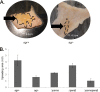
The human pathogen Staphylococcus aureus is renowned for the rapid colonization of contaminated wounds, medical implants, and food products. Nevertheless, little is known about the mechanisms that allow S. aureus to colonize the respective wet surfaces. The present studies were therefore aimed at identifying factors used by S. aureus cells to spread over wet surfaces, starting either from planktonic or biofilm-associated states. Through proteomics analyses we pinpoint phenol-soluble modulins (PSMs) as prime facilitators of the spreading process. To dissect the roles of the eight PSMs produced by S. aureus, these peptides were chemically synthesized and tested in spreading assays with different psm mutant strains. The results show that PSMα3 and PSMγ are the strongest facilitators of spreading both for planktonic cells and cells in catheter-associated biofilms. Compared to the six other PSMs of S. aureus, PSMα3 and PSMγ combine strong surfactant activities with a relatively low overall hydropathicity. Importantly, we show that PSM-mediated motility of S. aureus facilitates the rapid colonization of wet surfaces next to catheters and the colonization of fresh meat.
Figures
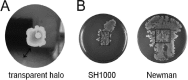


Similar articles
-
Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms.PLoS Pathog. 2012;8(6):e1002744. doi: 10.1371/journal.ppat.1002744. Epub 2012 Jun 7. PLoS Pathog. 2012. PMID: 22685403 Free PMC article.
-
Cross-talk between individual phenol-soluble modulins in Staphylococcus aureus biofilm enables rapid and efficient amyloid formation.Elife. 2020 Dec 1;9:e59776. doi: 10.7554/eLife.59776. Elife. 2020. PMID: 33259287 Free PMC article.
-
How Staphylococcus aureus biofilms develop their characteristic structure.Proc Natl Acad Sci U S A. 2012 Jan 24;109(4):1281-6. doi: 10.1073/pnas.1115006109. Epub 2012 Jan 9. Proc Natl Acad Sci U S A. 2012. PMID: 22232686 Free PMC article.
-
Phenol-soluble modulins.Int J Med Microbiol. 2014 Mar;304(2):164-9. doi: 10.1016/j.ijmm.2013.11.019. Epub 2013 Dec 1. Int J Med Microbiol. 2014. PMID: 24447915 Free PMC article. Review.
-
Phenol-soluble modulins and staphylococcal infection.Nat Rev Microbiol. 2013 Oct;11(10):667-73. doi: 10.1038/nrmicro3110. Epub 2013 Sep 10. Nat Rev Microbiol. 2013. PMID: 24018382 Free PMC article. Review.
Cited by
-
Pathophysiological Mechanisms of Staphylococcus Non-aureus Bone and Joint Infection: Interspecies Homogeneity and Specific Behavior of S. pseudintermedius.Front Microbiol. 2016 Jul 12;7:1063. doi: 10.3389/fmicb.2016.01063. eCollection 2016. Front Microbiol. 2016. PMID: 27462303 Free PMC article.
-
Modulation of Staphylococcus aureus Biofilm Matrix by Subinhibitory Concentrations of Clindamycin.Antimicrob Agents Chemother. 2016 Sep 23;60(10):5957-67. doi: 10.1128/AAC.00463-16. Print 2016 Oct. Antimicrob Agents Chemother. 2016. PMID: 27458233 Free PMC article.
-
4-Chloro-2-Isopropyl-5-Methylphenol Exhibits Antimicrobial and Adjuvant Activity against Methicillin-Resistant Staphylococcus aureus.J Microbiol Biotechnol. 2022 Jun 28;32(6):730-739. doi: 10.4014/jmb.2203.03054. Epub 2022 May 6. J Microbiol Biotechnol. 2022. PMID: 35586930 Free PMC article.
-
Multifunctional Amyloids in the Biology of Gram-Positive Bacteria.Microorganisms. 2020 Dec 17;8(12):2020. doi: 10.3390/microorganisms8122020. Microorganisms. 2020. PMID: 33348645 Free PMC article. Review.
-
Functional characteristics of the Staphylococcus aureus δ-toxin allelic variant G10S.Sci Rep. 2015 Dec 10;5:18023. doi: 10.1038/srep18023. Sci Rep. 2015. PMID: 26658455 Free PMC article.
References
-
- Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 - PubMed
-
- Peacock SJ, de Silva I, Lowy FD. 2001. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol. 9:605–610 - PubMed
-
- Ziebandt AK, Kusch H, Degner M, Jaglitz S, Sibbald MJ, Arends JP, Chlebowicz MA, Albrecht D, Pantucek R, Doskar J, Ziebuhr W, Broker BM, Hecker M, van Dijl JM, Engelmann S. 2010. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 10:1634–1644 - PubMed
-
- Dreisbach A, Hempel K, Buist G, Hecker M, Becher D, van Dijl JM. 2010. Profiling the surfaceome of Staphylococcus aureus. Proteomics 10:3082–3096 - PubMed
-
- Dreisbach A, van Dijl JM, Buist G. 2011. The cell surface proteome of Staphylococcus aureus. Proteomics 11:3154–3168 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

