Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels
- PMID: 23183982
- PMCID: PMC3568569
- DOI: 10.1128/AEM.02736-12
Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels
Abstract
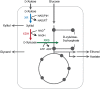
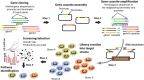
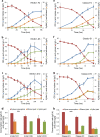
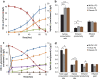
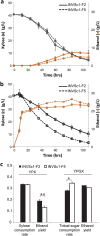
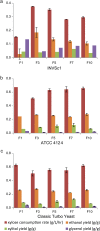
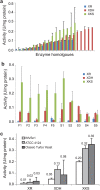
Balancing the flux of a heterologous metabolic pathway by tuning the expression and properties of the pathway enzymes is difficult, but it is critical to realizing the full potential of microbial biotechnology. One prominent example is the metabolic engineering of a Saccharomyces cerevisiae strain harboring a heterologous xylose-utilizing pathway for cellulosic-biofuel production, which remains a challenge even after decades of research. Here, we developed a combinatorial pathway-engineering approach to rapidly create a highly efficient xylose-utilizing pathway for ethanol production by exploring various combinations of enzyme homologues with different properties. A library of more than 8,000 xylose utilization pathways was generated using DNA assembler, followed by multitiered screening, which led to the identification of a number of strain-specific combinations of the enzymes for efficient conversion of xylose to ethanol. The balancing of metabolic flux through the xylose utilization pathway was demonstrated by a complete reversal of the major product from xylitol to ethanol with a similar yield and total by-product formation as low as 0.06 g/g xylose without compromising cell growth. The results also suggested that an optimal enzyme combination depends on not only the genotype/phenotype of the host strain, but also the sugar composition of the fermentation medium. This combinatorial approach should be applicable to any heterologous pathway and will be instrumental in the optimization of industrial production of value-added products.
Figures

Similar articles
-
Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective.Microb Cell Fact. 2017 May 11;16(1):82. doi: 10.1186/s12934-017-0694-9. Microb Cell Fact. 2017. PMID: 28494761 Free PMC article. Review.
-
Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae.Bioresour Technol. 2017 Mar;228:355-361. doi: 10.1016/j.biortech.2016.12.042. Epub 2016 Dec 18. Bioresour Technol. 2017. PMID: 28088640
-
Co-fermentation of cellobiose and xylose by mixed culture of recombinant Saccharomyces cerevisiae and kinetic modeling.PLoS One. 2018 Jun 25;13(6):e0199104. doi: 10.1371/journal.pone.0199104. eCollection 2018. PLoS One. 2018. PMID: 29940003 Free PMC article.
-
Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast.Nat Commun. 2013;4:2580. doi: 10.1038/ncomms3580. Nat Commun. 2013. PMID: 24105024
-
Recent progress in engineering yeast producers of cellulosic ethanol.FEMS Yeast Res. 2025 Jan 30;25:foaf035. doi: 10.1093/femsyr/foaf035. FEMS Yeast Res. 2025. PMID: 40613832 Free PMC article. Review.
Cited by
-
Characterization of Thermotoga neapolitana Alcohol Dehydrogenases in the Ethanol Fermentation Pathway.Biology (Basel). 2022 Sep 5;11(9):1318. doi: 10.3390/biology11091318. Biology (Basel). 2022. PMID: 36138797 Free PMC article.
-
Systematic Optimization of Protein Secretory Pathways in Saccharomyces cerevisiae to Increase Expression of Hepatitis B Small Antigen.Front Microbiol. 2017 May 16;8:875. doi: 10.3389/fmicb.2017.00875. eCollection 2017. Front Microbiol. 2017. PMID: 28559891 Free PMC article.
-
Dynamic modulation of enzyme activity by synthetic CRISPR-Cas6 endonucleases.Nat Chem Biol. 2022 May;18(5):492-500. doi: 10.1038/s41589-022-01005-7. Epub 2022 Apr 25. Nat Chem Biol. 2022. PMID: 35468950
-
DNA assembly techniques for next-generation combinatorial biosynthesis of natural products.J Ind Microbiol Biotechnol. 2014 Feb;41(2):469-77. doi: 10.1007/s10295-013-1358-3. Epub 2013 Oct 15. J Ind Microbiol Biotechnol. 2014. PMID: 24127070 Free PMC article. Review.
-
Investigating xylose metabolism in recombinant Saccharomyces cerevisiae via 13C metabolic flux analysis.Microb Cell Fact. 2013 Nov 18;12:114. doi: 10.1186/1475-2859-12-114. Microb Cell Fact. 2013. PMID: 24245823 Free PMC article.
References
-
- Ramon A, Smith HO. 2011. Single-step linker-based combinatorial assembly of promoter and gene cassettes for pathway engineering. Biotechnol. Lett. 33:549–555 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

