Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation
- PMID: 23184059
- PMCID: PMC3515759
- DOI: 10.1038/cr.2012.160
Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation
Erratum in
-
Transcriptional activation of transposable elements in mouse zygotes is independent of Tet3-mediated 5-methylcytosine oxidation.Cell Res. 2018 Feb;28(2):261. doi: 10.1038/cr.2018.4. Cell Res. 2018. PMID: 29508854 Free PMC article.
Abstract
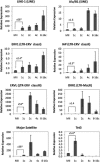
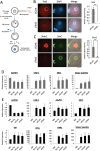
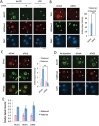
The methylation state of the paternal genome is rapidly reprogrammed shortly after fertilization. Recent studies have revealed that loss of 5-methylcytosine (5mC) in zygotes correlates with appearance of 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). This process is mediated by Tet3 and the 5mC oxidation products generated in zygotes are gradually lost during preimplantation development through a replication-dependent dilution process. Despite these findings, the biological significance of Tet3-mediated oxidation of 5mC to 5hmC/5fC/5caC in zygotes is unknown. DNA methylation plays an important role in silencing gene expression including the repression of transposable elements (TEs). Given that the activation of TEs during preimplantation development correlates with loss of DNA methylation, it is believed that paternal DNA demethylation may have an important role in TE activation. Here we examined this hypothesis and found that Tet3-mediated 5mC oxidation does not have a significant contribution to TE activation. We show that the expression of LINE-1 (long interspersed nucleotide element 1) and ERVL (endogenous retroviruses class III) are activated from both paternal and maternal genomes in zygotes. Inhibition of 5mC oxidation by siRNA-mediated depletion of Tet3 affected neither TE activation, nor global transcription in zygotes. Thus, our study provides the first evidence demonstrating that activation of both TEs and global transcription in zygotes are independent of Tet3-mediated 5mC oxidation.
Figures

Similar articles
-
Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes.Cell Stem Cell. 2014 Oct 2;15(4):459-471. doi: 10.1016/j.stem.2014.09.002. Cell Stem Cell. 2014. PMID: 25280220 Free PMC article.
-
The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes.Nature. 2011 Sep 4;477(7366):606-10. doi: 10.1038/nature10443. Nature. 2011. PMID: 21892189
-
Haploinsufficiency, but not defective paternal 5mC oxidation, accounts for the developmental defects of maternal Tet3 knockouts.Cell Rep. 2015 Feb 3;10(4):463-70. doi: 10.1016/j.celrep.2014.12.049. Epub 2015 Jan 29. Cell Rep. 2015. PMID: 25640176 Free PMC article.
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
-
Structure and Function of TET Enzymes.Adv Exp Med Biol. 2016;945:275-302. doi: 10.1007/978-3-319-43624-1_12. Adv Exp Med Biol. 2016. PMID: 27826843 Review.
Cited by
-
Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes.Cell Stem Cell. 2014 Oct 2;15(4):459-471. doi: 10.1016/j.stem.2014.09.002. Cell Stem Cell. 2014. PMID: 25280220 Free PMC article.
-
TET-mediated active DNA demethylation: mechanism, function and beyond.Nat Rev Genet. 2017 Sep;18(9):517-534. doi: 10.1038/nrg.2017.33. Epub 2017 May 30. Nat Rev Genet. 2017. PMID: 28555658 Review.
-
Mechanisms underlying low mutation rates in mammalian oocytes and preimplantation embryos.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf760. doi: 10.1093/nar/gkaf760. Nucleic Acids Res. 2025. PMID: 40795959 Free PMC article. Review.
-
Delineation of a Human Mendelian Disorder of the DNA Demethylation Machinery: TET3 Deficiency.Am J Hum Genet. 2020 Feb 6;106(2):234-245. doi: 10.1016/j.ajhg.2019.12.007. Epub 2020 Jan 9. Am J Hum Genet. 2020. PMID: 31928709 Free PMC article.
-
LINEs in mice: features, families, and potential roles in early development.Chromosoma. 2016 Mar;125(1):29-39. doi: 10.1007/s00412-015-0520-2. Epub 2015 May 16. Chromosoma. 2016. PMID: 25975894 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

