Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma
- PMID: 23184418
- PMCID: PMC3580007
- DOI: 10.1007/s00401-012-1070-9
Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma
Abstract
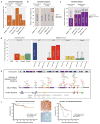
Recent sequencing efforts have described the mutational landscape of the pediatric brain tumor medulloblastoma. Although MLL2 is among the most frequent somatic single nucleotide variants (SNV), the clinical and biological significance of these mutations remains uncharacterized. Through targeted re-sequencing, we identified mutations of MLL2 in 8 % (14/175) of MBs, the majority of which were loss of function. Notably, we also report mutations affecting the MLL2-binding partner KDM6A, in 4 % (7/175) of tumors. While MLL2 mutations were independent of age, gender, histological subtype, M-stage or molecular subgroup, KDM6A mutations were most commonly identified in Group 4 MBs, and were mutually exclusive with MLL2 mutations. Immunohistochemical staining for H3K4me3 and H3K27me3, the chromatin effectors of MLL2 and KDM6A activity, respectively, demonstrated alterations of the histone code in 24 % (53/220) of MBs across all subgroups. Correlating these MLL2- and KDM6A-driven histone marks with prognosis, we identified populations of MB with improved (K4+/K27-) and dismal (K4-/K27-) outcomes, observed primarily within Group 3 and 4 MBs. Group 3 and 4 MBs demonstrate somatic copy number aberrations, and transcriptional profiles that converge on modifiers of H3K27-methylation (EZH2, KDM6A, KDM6B), leading to silencing of PRC2-target genes. As PRC2-mediated aberrant methylation of H3K27 has recently been targeted for therapy in other diseases, it represents an actionable target for a substantial percentage of medulloblastoma patients with aggressive forms of the disease.
Figures



References
-
- Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15(12):3970–3977. - PubMed
-
- Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, et al. Antitumor effects of a combined 5-aza-2′ deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res. 2009;69(3):887–895. - PubMed
-
- Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23(31):7951–7957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

