Using QIIME to analyze 16S rRNA gene sequences from microbial communities
- PMID: 23184592
- PMCID: PMC4477843
- DOI: 10.1002/9780471729259.mc01e05s27
Using QIIME to analyze 16S rRNA gene sequences from microbial communities
Abstract
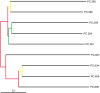
QIIME (canonically pronounced "chime") is a software application that performs microbial community analysis. It is an acronym for Quantitative Insights Into Microbial Ecology, and has been used to analyze and interpret nucleic acid sequence data from fungal, viral, bacterial, and archaeal communities. The following protocols describe how to install QIIME on a single computer and use it to analyze microbial 16S sequence data from nine distinct microbial communities.
© 2012 by John Wiley & Sons, Inc.
Figures



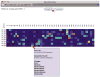



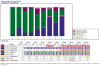
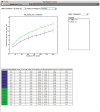
References
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P MetaHIT Consortium. Enterotypes of the human gut microbiome. Nature. 2011;474:666. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG4872/HG/NHGRI NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U01 HG004866/HG/NHGRI NIH HHS/United States
- GM65103/GM/NIGMS NIH HHS/United States
- R01 HG004872/HG/NHGRI NIH HHS/United States
- GM8759/GM/NIGMS NIH HHS/United States
- HG4866/HG/NHGRI NIH HHS/United States
- LM9451/LM/NLM NIH HHS/United States
- DK83981/DK/NIDDK NIH HHS/United States
- UH2 DK083981/DK/NIDDK NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- UH3 DK083981/DK/NIDDK NIH HHS/United States
- T15 LM009451/LM/NLM NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
- T32 GM065103/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

