Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea
- PMID: 23184964
- PMCID: PMC3528564
- DOI: 10.1073/pnas.1209119109
Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea
Abstract
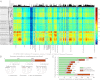
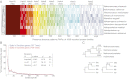
Archaebacterial halophiles (Haloarchaea) are oxygen-respiring heterotrophs that derive from methanogens--strictly anaerobic, hydrogen-dependent autotrophs. Haloarchaeal genomes are known to have acquired, via lateral gene transfer (LGT), several genes from eubacteria, but it is yet unknown how many genes the Haloarchaea acquired in total and, more importantly, whether independent haloarchaeal lineages acquired their genes in parallel, or as a single acquisition at the origin of the group. Here we have studied 10 haloarchaeal and 1,143 reference genomes and have identified 1,089 haloarchaeal gene families that were acquired by a methanogenic recipient from eubacteria. The data suggest that these genes were acquired in the haloarchaeal common ancestor, not in parallel in independent haloarchaeal lineages, nor in the common ancestor of haloarchaeans and methanosarcinales. The 1,089 acquisitions include genes for catabolic carbon metabolism, membrane transporters, menaquinone biosynthesis, and complexes I-IV of the eubacterial respiratory chain that functions in the haloarchaeal membrane consisting of diphytanyl isoprene ether lipids. LGT on a massive scale transformed a strictly anaerobic, chemolithoautotrophic methanogen into the heterotrophic, oxygen-respiring, and bacteriorhodopsin-photosynthetic haloarchaeal common ancestor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Metabolic bacterial genes and the construction of high-level composite lineages of life.Trends Ecol Evol. 2015 Mar;30(3):127-9. doi: 10.1016/j.tree.2015.01.001. Epub 2015 Jan 16. Trends Ecol Evol. 2015. PMID: 25601290 Free PMC article.
References
-
- Oren A. Life at high salt concentrations. The Prokaryotes. 2006;3:263–282.
-
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6(3):245–252. - PubMed
-
- Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6(8):579–591. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

