Alternative bacterial two-component small heat shock protein systems
- PMID: 23184973
- PMCID: PMC3528540
- DOI: 10.1073/pnas.1209565109
Alternative bacterial two-component small heat shock protein systems
Abstract
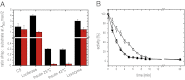
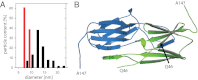
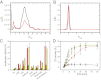
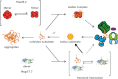
Small heat shock proteins (sHsps) are molecular chaperones that prevent the aggregation of nonnative proteins. The sHsps investigated to date mostly form large, oligomeric complexes. The typical bacterial scenario seemed to be a two-component sHsps system of two homologous sHsps, such as the Escherichia coli sHsps IbpA and IbpB. With a view to expand our knowledge on bacterial sHsps, we analyzed the sHsp system of the bacterium Deinococcus radiodurans, which is resistant against various stress conditions. D. radiodurans encodes two sHsps, termed Hsp17.7 and Hsp20.2. Surprisingly, Hsp17.7 forms only chaperone active dimers, although its crystal structure reveals the typical α-crystallin fold. In contrast, Hsp20.2 is predominantly a 36mer that dissociates into smaller oligomeric assemblies that bind substrate proteins stably. Whereas Hsp20.2 cooperates with the ATP-dependent bacterial chaperones in their refolding, Hsp17.7 keeps substrates in a refolding-competent state by transient interactions. In summary, we show that these two sHsps are strikingly different in their quaternary structures and chaperone properties, defining a second type of bacterial two-component sHsp system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12(10):842–846. - PubMed
-
- Richter K, Haslbeck M, Buchner J. The heat shock response: Life on the verge of death. Mol Cell. 2010;40(2):253–266. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

