Ezrin interacts with the SARS coronavirus Spike protein and restrains infection at the entry stage
- PMID: 23185364
- PMCID: PMC3504146
- DOI: 10.1371/journal.pone.0049566
Ezrin interacts with the SARS coronavirus Spike protein and restrains infection at the entry stage
Abstract
Background: Entry of Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and its envelope fusion with host cell membrane are controlled by a series of complex molecular mechanisms, largely dependent on the viral envelope glycoprotein Spike (S). There are still many unknowns on the implication of cellular factors that regulate the entry process.
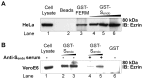
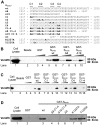
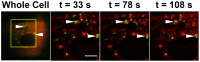
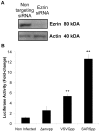
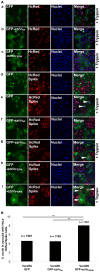
Methodology/principal findings: We performed a yeast two-hybrid screen using as bait the carboxy-terminal endodomain of S, which faces the cytosol during and after opening of the fusion pore at early stages of the virus life cycle. Here we show that the ezrin membrane-actin linker interacts with S endodomain through the F1 lobe of its FERM domain and that both the eight carboxy-terminal amino-acids and a membrane-proximal cysteine cluster of S endodomain are important for this interaction in vitro. Interestingly, we found that ezrin is present at the site of entry of S-pseudotyped lentiviral particles in Vero E6 cells. Targeting ezrin function by small interfering RNA increased S-mediated entry of pseudotyped particles in epithelial cells. Furthermore, deletion of the eight carboxy-terminal amino acids of S enhanced S-pseudotyped particles infection. Expression of the ezrin dominant negative FERM domain enhanced cell susceptibility to infection by SARS-CoV and S-pseudotyped particles and potentiated S-dependent membrane fusion.
Conclusions/significance: Ezrin interacts with SARS-CoV S endodomain and limits virus entry and fusion. Our data present a novel mechanism involving a cellular factor in the regulation of S-dependent early events of infection.
Conflict of interest statement
Figures

Similar articles
-
Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion.Virology. 2005 Oct 25;341(2):215-30. doi: 10.1016/j.virol.2005.06.046. Epub 2005 Aug 15. Virology. 2005. PMID: 16099010 Free PMC article.
-
Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion.Virology. 2007 Apr 10;360(2):264-74. doi: 10.1016/j.virol.2006.10.034. Epub 2006 Nov 28. Virology. 2007. PMID: 17134730 Free PMC article.
-
Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor.J Virol. 2004 May;78(9):4552-60. doi: 10.1128/jvi.78.9.4552-4560.2004. J Virol. 2004. PMID: 15078936 Free PMC article.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
-
Insights from the association of SARS-CoV S-protein with its receptor, ACE2.Adv Exp Med Biol. 2006;581:209-18. doi: 10.1007/978-0-387-33012-9_36. Adv Exp Med Biol. 2006. PMID: 17037532 Free PMC article. Review. No abstract available.
Cited by
-
A comprehensive review of COVID-19 characteristics.Biol Proced Online. 2020 Aug 4;22:19. doi: 10.1186/s12575-020-00128-2. eCollection 2020. Biol Proced Online. 2020. PMID: 32774178 Free PMC article. Review.
-
Cell Entry of Animal Coronaviruses.Viruses. 2021 Oct 1;13(10):1977. doi: 10.3390/v13101977. Viruses. 2021. PMID: 34696406 Free PMC article. Review.
-
Murine Leukemia Virus (MLV)-based Coronavirus Spike-pseudotyped Particle Production and Infection.Bio Protoc. 2016 Dec 5;6(23):e2035. doi: 10.21769/BioProtoc.2035. Bio Protoc. 2016. PMID: 28018942 Free PMC article.
-
Designing spike protein (S-Protein) based multi-epitope peptide vaccine against SARS COVID-19 by immunoinformatics.Heliyon. 2020 Nov;6(11):e05528. doi: 10.1016/j.heliyon.2020.e05528. Epub 2020 Nov 16. Heliyon. 2020. PMID: 33225084 Free PMC article.
-
Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications.Sci Signal. 2021 Jan 12;14(665):eabd0334. doi: 10.1126/scisignal.abd0334. Sci Signal. 2021. PMID: 33436497 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

