Adaptation for protein synthesis efficiency in a naturally occurring self-regulating operon
- PMID: 23185406
- PMCID: PMC3502259
- DOI: 10.1371/journal.pone.0049678
Adaptation for protein synthesis efficiency in a naturally occurring self-regulating operon
Abstract
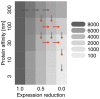
The korAB operon in RK2 plasmids is a beautiful natural example of a negatively and cooperatively self-regulating operon. It has been particularly well characterized both experimentally and with mathematical models. We have carried out a detailed investigation of the role of the regulatory mechanism using a biologically grounded mechanistic multi-scale stochastic model that includes plasmid gene regulation and replication in the context of host growth and cell division. We use the model to compare four hypotheses for the action of the regulatory mechanism: increased robustness to extrinsic factors, decreased protein fluctuations, faster response-time of the operon and reduced host burden through improved efficiency of protein production. We find that the strongest impact of all elements of the regulatory architecture is on improving the efficiency of protein synthesis by reduction in the number of mRNA molecules needed to be produced, leading to a greater than ten-fold reduction in host energy required to express these plasmid proteins. A smaller but still significant role is seen for speeding response times, but this is not materially improved by the cooperativity. The self-regulating mechanisms have the least impact on protein fluctuations and robustness. While reduction of host burden is evident in a plasmid context, negative self-regulation is a widely seen motif for chromosomal genes. We propose that an important evolutionary driver for negatively self-regulated genes is to improve the efficiency of protein synthesis.
Conflict of interest statement
Figures


References
-
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, et al. (2002) Network motifs: simple building blocks of complex networks. Science (New York, NY) 298: 824–827. - PubMed
-
- Becskei A, Serrano L (2000) Engineering stability in gene networks by autoregulation. Nature 405: 590–593. - PubMed
-
- Rosenfeld N, Elowitz MB, Alon U (2002) Negative Autoregulation Speeds the Response Times of Transcription Networks. Journal of Molecular Biology 323: 785–793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

