Genetic variation within native populations of endemic silkmoth Antheraea assamensis (Helfer) from Northeast India indicates need for in situ conservation
- PMID: 23185503
- PMCID: PMC3503872
- DOI: 10.1371/journal.pone.0049972
Genetic variation within native populations of endemic silkmoth Antheraea assamensis (Helfer) from Northeast India indicates need for in situ conservation
Abstract
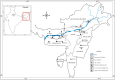
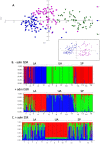
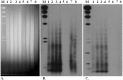
A. assamensis is a phytophagous Lepidoptera from Northeast India reared on host trees of Lauraceae family for its characteristic cocoon silk. Source of these cocoons are domesticated farm stocks that crash frequently and/or wild insect populations that provide new cultures. The need to reduce dependence on wild populations for cocoons necessitates assessment of genetic diversity in cultivated and wild populations. Molecular markers based on PCR of Inter-simple sequence repeats (ISSR) and simple sequence repeats (SSR) were used with four populations of wild insects and eleven populations of cultivated insects. Wild populations had high genetic diversity estimates (H(i) = 0.25; H(S) = 0.28; H(E) = 0.42) and at least one population contained private alleles. Both marker systems indicated that genetic variability within populations examined was significantly high. Among cultivated populations, insects of the Upper Assam region (H(i) = 0.19; H(S) = 0.18; H(E) = 0) were genetically distinct (F(ST) = 0.38 with both marker systems) from insects of Lower Assam (H(i) =0.24; H(S) =0.25; H(E) = 0.3). Sequencing of polymorphic amplicons suggested transposition as a mechanism for maintaining genomic diversity. Implications for conservation of native populations in the wild and preserving in-farm diversity are discussed.
Conflict of interest statement
Figures

Similar articles
-
Genetic diversity and population structure of Indian golden silkmoth (Antheraea assama).PLoS One. 2012;7(8):e43716. doi: 10.1371/journal.pone.0043716. Epub 2012 Aug 28. PLoS One. 2012. PMID: 22952746 Free PMC article.
-
De novo transcriptome of the muga silkworm, Antheraea assamensis (Helfer).Gene. 2017 May 5;611:54-65. doi: 10.1016/j.gene.2017.02.021. Epub 2017 Feb 17. Gene. 2017. PMID: 28216038
-
Genetic variability and genetic structure of wild and semi-domestic populations of tasar silkworm (Antheraea mylitta ) ecorace Daba as revealed through ISSR markers.Genetica. 2005 Nov;125(2-3):173-83. doi: 10.1007/s10709-005-7002-z. Genetica. 2005. PMID: 16247690
-
Genome sequencing and assembly of Indian golden silkmoth, Antheraea assamensis Helfer (Saturniidae, Lepidoptera).Genomics. 2024 May;116(3):110841. doi: 10.1016/j.ygeno.2024.110841. Epub 2024 Apr 9. Genomics. 2024. PMID: 38599255
-
The mitochondrial genome of Muga silkworm (Antheraea assamensis) and its comparative analysis with other lepidopteran insects.PLoS One. 2017 Nov 15;12(11):e0188077. doi: 10.1371/journal.pone.0188077. eCollection 2017. PLoS One. 2017. PMID: 29141006 Free PMC article.
Cited by
-
Assessment of various genetic components through NCD-I and NCD-III designs of biparental mating in opium poppy.J Genet. 2019 Mar;98:27. J Genet. 2019. PMID: 30945678
-
Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis.PLoS One. 2013 Dec 12;8(12):e83253. doi: 10.1371/journal.pone.0083253. eCollection 2013. PLoS One. 2013. PMID: 24349475 Free PMC article.
-
Genetic analysis of Indian tasar silkmoth (Antheraea mylitta) populations.Sci Rep. 2015 Oct 29;5:15728. doi: 10.1038/srep15728. Sci Rep. 2015. PMID: 26510465 Free PMC article.
-
Genetic diversity analysis of Saccharosydne procerus in Hunan region, China.Mol Biol Rep. 2024 Jul 31;51(1):878. doi: 10.1007/s11033-024-09770-5. Mol Biol Rep. 2024. PMID: 39083078
-
High genetic diversity and structured populations of the oriental fruit moth in its range of origin.PLoS One. 2013 Nov 4;8(11):e78476. doi: 10.1371/journal.pone.0078476. eCollection 2013. PLoS One. 2013. PMID: 24265692 Free PMC article.
References
-
- Hugon T (1837) Remarks on the silkworms and silks of Assam. J Asiatic Soc Bengal 6: 21–38.
-
- Chowdhury SN (2001) Sericulture and weaving (an overview). Designer Graphics Press Dibrugarh, Assam, India.
-
- Good IL, Kenoyer JM, Meadow RH (2009) New evidence for early silk in the Indus civilization. Archaeometry 50: 457–466.
-
- Bindroo BB, Singh NT, Sahu AK, Chakravorty R (2006) Muga silkworm host plants. Indian Silk 44: 13–17.
-
- Singh KC and Chakravorty R (2006) Seri-biodiversity of North-Eastern India-an update. In JP Handique and MC Kalita (eds.) Biodiversity, Conservation and Future concern. Gauhati University, Gauhati, India. 8–19.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

