Thermal, chemical and pH induced denaturation of a multimeric β-galactosidase reveals multiple unfolding pathways
- PMID: 23185611
- PMCID: PMC3503960
- DOI: 10.1371/journal.pone.0050380
Thermal, chemical and pH induced denaturation of a multimeric β-galactosidase reveals multiple unfolding pathways
Abstract
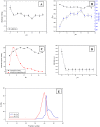
Background: In this case study, we analysed the properties of unfolded states and pathways leading to complete denaturation of a multimeric chick pea β-galactosidase (CpGAL), as obtained from treatment with guanidium hydrochloride, urea, elevated temperature and extreme pH.
Methodology/principal findings: CpGAL, a heterodimeric protein with native molecular mass of 85 kDa, belongs to α+β class of protein. The conformational stability and thermodynamic parameters of CpGAL unfolding in different states were estimated and interpreted using circular dichroism and fluorescence spectroscopic measurements. The enzyme was found to be structurally and functionally stable in the entire pH range and upto 50 °C temperature. Further increase in temperature induces unfolding followed by aggregation. Chemical induced denaturation was found to be cooperative and transitions were irreversible, non-coincidental and sigmoidal. Free energy of protein unfolding (ΔG(0)) and unfolding constant (K(obs)) were also calculated for chemically denatured CpGAL.
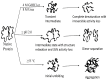
Significance: The protein seems to use different pathways for unfolding in different environments and is a classical example of how the environment dictates the path a protein might take to fold while its amino acid sequence only defines its final three-dimensional conformation. The knowledge accumulated could be of immense biotechnological significance as well.
Conflict of interest statement
Figures



Similar articles
-
Heat, Acid and Chemically Induced Unfolding Pathways, Conformational Stability and Structure-Function Relationship in Wheat α-Amylase.PLoS One. 2015 Jun 8;10(6):e0129203. doi: 10.1371/journal.pone.0129203. eCollection 2015. PLoS One. 2015. PMID: 26053142 Free PMC article.
-
Conformational plasticity of cryptolepain: accumulation of partially unfolded states in denaturants induced equilibrium unfolding.J Biotechnol. 2007 Sep 30;131(4):404-17. doi: 10.1016/j.jbiotec.2007.08.006. Epub 2007 Aug 7. J Biotechnol. 2007. PMID: 17825936
-
Accumulation of partly folded states in the equilibrium unfolding of ervatamin A: spectroscopic description of the native, intermediate, and unfolded states.Biochimie. 2007 Nov;89(11):1416-24. doi: 10.1016/j.biochi.2007.06.004. Epub 2007 Jun 8. Biochimie. 2007. PMID: 17658212
-
An "Onion-like" Model of Protein Unfolding: Collective versus Site Specific Approaches.Chemphyschem. 2022 Jan 5;23(1):e202100520. doi: 10.1002/cphc.202100520. Epub 2021 Oct 13. Chemphyschem. 2022. PMID: 34549492 Review.
-
Conformational Stability and Denaturation Processes of Proteins Investigated by Electrophoresis under Extreme Conditions.Molecules. 2022 Oct 13;27(20):6861. doi: 10.3390/molecules27206861. Molecules. 2022. PMID: 36296453 Free PMC article. Review.
Cited by
-
Investigating thermal stability based on the structural changes of lactase enzyme by several orthogonal methods.Biotechnol Rep (Amst). 2021 May 26;30:e00637. doi: 10.1016/j.btre.2021.e00637. eCollection 2021 Jun. Biotechnol Rep (Amst). 2021. PMID: 34136367 Free PMC article.
-
Atomic Force Microscopy Study of the Temperature and Storage Duration Dependencies of Horseradish Peroxidase Oligomeric State.Biomedicines. 2022 Oct 20;10(10):2645. doi: 10.3390/biomedicines10102645. Biomedicines. 2022. PMID: 36289907 Free PMC article.
-
Identification and characterisation of moderately thermostable diisobutyl phthalate degrading esterase from a Great Artesian Basin Bacillus velezensis NP05.Biotechnol Rep (Amst). 2024 Apr 4;42:e00840. doi: 10.1016/j.btre.2024.e00840. eCollection 2024 Jun. Biotechnol Rep (Amst). 2024. PMID: 38645886 Free PMC article.
-
Following Structural Changes by Thermal Denaturation Using Trapped Ion Mobility Spectrometry-Mass Spectrometry.J Phys Chem B. 2020 Jul 23;124(29):6257-6265. doi: 10.1021/acs.jpcb.0c04276. Epub 2020 Jul 14. J Phys Chem B. 2020. PMID: 32560586 Free PMC article.
-
Heat, Acid and Chemically Induced Unfolding Pathways, Conformational Stability and Structure-Function Relationship in Wheat α-Amylase.PLoS One. 2015 Jun 8;10(6):e0129203. doi: 10.1371/journal.pone.0129203. eCollection 2015. PLoS One. 2015. PMID: 26053142 Free PMC article.
References
-
- Nichtl A, Buchner J, Jaenicke R, Rudolph R, Scheibel T (1998) Folding and association of β-galactosidase. J Mol Biol 282: 1083–1091. - PubMed
-
- Radford SE (2000) Protein folding: progress made and promises ahead. Trends Biochem Sci 25: 611–618. - PubMed
-
- Jagannadham MV, Dubey VK (2003) Differences in the unfolding of procerain induced by pH, guanidine hydrochloride, urea, and temperature. Biochemistry 42: 12287–12297. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

