Chitinase mRNA levels by quantitative PCR using the single standard DNA: acidic mammalian chitinase is a major transcript in the mouse stomach
- PMID: 23185612
- PMCID: PMC3503932
- DOI: 10.1371/journal.pone.0050381
Chitinase mRNA levels by quantitative PCR using the single standard DNA: acidic mammalian chitinase is a major transcript in the mouse stomach
Abstract
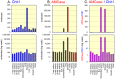
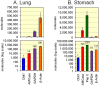
Chitinases hydrolyze the β-1-4 glycosidic bonds of chitin, a major structural component of fungi, crustaceans and insects. Although mammals do not produce chitin or its synthase, they express two active chitinases, chitotriosidase (Chit1) and acidic mammalian chitinase (AMCase). These mammalian chitinases have attracted considerable attention due to their increased expression in individuals with a number of pathological conditions, including Gaucher disease, Alzheimer's disease and asthma. However, the contribution of these enzymes to the pathophysiology of these diseases remains to be determined. The quantification of the Chit1 and AMCase mRNA levels and the comparison of those levels with the levels of well-known reference genes can generate useful and biomedically relevant information. In the beginning, we established a quantitative real-time PCR system that uses standard DNA produced by ligating the cDNA fragments of the target genes. This system enabled us to quantify and compare the expression levels of the chitinases and the reference genes on the same scale. We found that AMCase mRNA is synthesized at extraordinarily high levels in the mouse stomach. The level of this mRNA in the mouse stomach was 7- to 10-fold higher than the levels of the housekeeping genes and was comparable to that the level of the mRNA for pepsinogen C (progastricsin), a major component of the gastric mucosa. Thus, AMCase mRNA is a major transcript in mouse stomach, suggesting that AMCase functions as a digestive enzyme that breaks down polymeric chitin and as part of the host defense against chitin-containing pathogens in the gastric contents. Our methodology is applicable to the quantification of mRNAs for multiple genes across multiple specimens using the same scale.
Conflict of interest statement
Figures
Similar articles
-
Quantification of Chitinase mRNA Levels in Human and Mouse Tissues by Real-Time PCR: Species-Specific Expression of Acidic Mammalian Chitinase in Stomach Tissues.PLoS One. 2013 Jun 27;8(6):e67399. doi: 10.1371/journal.pone.0067399. Print 2013. PLoS One. 2013. PMID: 23826286 Free PMC article.
-
Protease resistance of porcine acidic mammalian chitinase under gastrointestinal conditions implies that chitin-containing organisms can be sustainable dietary resources.Sci Rep. 2017 Oct 11;7(1):12963. doi: 10.1038/s41598-017-13526-6. Sci Rep. 2017. PMID: 29021549 Free PMC article.
-
Acidic mammalian chitinase is a proteases-resistant glycosidase in mouse digestive system.Sci Rep. 2016 Nov 24;6:37756. doi: 10.1038/srep37756. Sci Rep. 2016. PMID: 27883045 Free PMC article.
-
Human Chitinases: Structure, Function, and Inhibitor Discovery.Adv Exp Med Biol. 2019;1142:221-251. doi: 10.1007/978-981-13-7318-3_11. Adv Exp Med Biol. 2019. PMID: 31102249 Review.
-
The biology of the Gaucher cell: the cradle of human chitinases.Int Rev Cytol. 2006;252:71-128. doi: 10.1016/S0074-7696(06)52001-7. Int Rev Cytol. 2006. PMID: 16984816 Review.
Cited by
-
Loss and Gain of Human Acidic Mammalian Chitinase Activity by Nonsynonymous SNPs.Mol Biol Evol. 2016 Dec;33(12):3183-3193. doi: 10.1093/molbev/msw198. Epub 2016 Oct 4. Mol Biol Evol. 2016. PMID: 27702777 Free PMC article.
-
Protein A-mouse acidic mammalian chitinase-V5-His expressed in periplasmic space of Escherichia coli possesses chitinase functions comparable to CHO-expressed protein.PLoS One. 2013 Nov 11;8(11):e78669. doi: 10.1371/journal.pone.0078669. eCollection 2013. PLoS One. 2013. PMID: 24244337 Free PMC article.
-
Gastric and intestinal proteases resistance of chicken acidic chitinase nominates chitin-containing organisms for alternative whole edible diets for poultry.Sci Rep. 2017 Jul 27;7(1):6662. doi: 10.1038/s41598-017-07146-3. Sci Rep. 2017. PMID: 28751762 Free PMC article.
-
Quantitative Real-Time PCR Analysis of YKL-40 and Its Comparison with Mammalian Chitinase mRNAs in Normal Human Tissues Using a Single Standard DNA.Int J Mol Sci. 2015 Apr 30;16(5):9922-35. doi: 10.3390/ijms16059922. Int J Mol Sci. 2015. PMID: 25941933 Free PMC article.
-
High expression of acidic chitinase and chitin digestibility in the stomach of common marmoset (Callithrix jacchus), an insectivorous nonhuman primate.Sci Rep. 2019 Jan 17;9(1):159. doi: 10.1038/s41598-018-36477-y. Sci Rep. 2019. PMID: 30655565 Free PMC article.
References
-
- Bussink AP, van Eijk M, Renkema GH, Aerts JM, Boot RG (2006) The biology of the Gaucher cell: the cradle of human chitinases. Int Rev Cytol 252: 71–128. - PubMed
-
- Renkema GH, Boot RG, Muijsers AO, Donker-Koopman WE, Aerts JM (1995) Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J Biol Chem 270: 2198–2202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

