Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells
- PMID: 23187807
- PMCID: PMC3562305
- DOI: 10.4161/cc.22871
Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells
Abstract
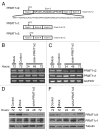
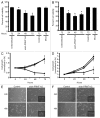
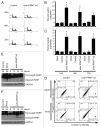
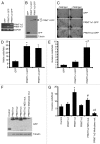
Protein arginine methylation is catalyzed by protein arginine methyltransferases (PRMTs) and plays an important role in many cellular processes. Aberrant PRMT expression has been observed in several common cancer types; however, their precise contribution to the cell transformation process is not well understood. We previously reported that the PRMT1 gene generates several alternatively spliced isoforms, and our initial biochemical characterization of these isoforms revealed that they exhibit distinct substrate specificity and subcellular localization. We focus here on the PRMT1v2 isoform, which is the only predominantly cytoplasmic isoform, and we have found that its relative expression is increased in breast cancer cell lines and tumors. Specific depletion of PRMT1v2 using RNA interference caused a significant decrease in cancer cell survival due to an induction of apoptosis. Furthermore, depletion of PRMT1v2 in an aggressive cancer cell line significantly decreased cell invasion. We also demonstrate that PRMT1v2 overexpression in a non-aggressive cancer cell line was sufficient to render them more invasive. Importantly, this novel activity is specific to PRMT1v2, as overexpression of other isoforms did not enhance invasion. Moreover, this activity requires both proper subcellular localization and methylase activity. Lastly, PRMT1v2 overexpression altered cell morphology and reduced cell-cell adhesion, a phenomenon that we convincingly linked with reduced β-catenin protein expression. Overall, we demonstrate a specific role for PRMT1v2 in breast cancer cell survival and invasion, underscoring the importance of identifying and characterizing the distinct functional differences between PRMT1 isoforms.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
