Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes
- PMID: 23188510
- PMCID: PMC3509411
- DOI: 10.1128/mBio.00333-12
Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes
Abstract
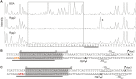
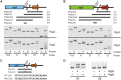
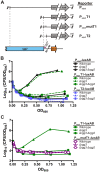
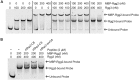
Recent studies have established the fact that multiple members of the Rgg family of transcriptional regulators serve as key components of quorum sensing (QS) pathways that utilize peptides as intercellular signaling molecules. We previously described a novel QS system in Streptococcus pyogenes which utilizes two Rgg-family regulators (Rgg2 and Rgg3) that respond to neighboring signaling peptides (SHP2 and SHP3) to control gene expression and biofilm formation. We have shown that Rgg2 is a transcriptional activator of target genes, whereas Rgg3 represses expression of these genes, and that SHPs function to activate the QS system. The mechanisms by which Rgg proteins regulate both QS-dependent and QS-independent processes remain poorly defined; thus, we sought to further elucidate how Rgg2 and Rgg3 mediate gene regulation. Here we provide evidence that S. pyogenes employs a unique mechanism of direct competition between the antagonistic, peptide-responsive proteins Rgg2 and Rgg3 for binding at target promoters. The highly conserved, shared binding sites for Rgg2 and Rgg3 are located proximal to the -35 nucleotide in the target promoters, and the direct competition between the two regulators results in concentration-dependent, exclusive occupation of the target promoters that can be skewed in favor of Rgg2 in vitro by the presence of SHP. These results suggest that exclusionary binding of target promoters by Rgg3 may prevent Rgg2 binding under SHP-limiting conditions, thereby preventing premature induction of the quorum sensing circuit.
Importance: Rgg-family transcriptional regulators are widespread among low-G+C Gram-positive bacteria and in many cases contribute to bacterial physiology and virulence. Only recently was it discovered that several Rgg proteins function in cell-to-cell communication (quorum sensing [QS]) via direct interaction with signaling peptides. The mechanism(s) by which Rgg proteins mediate regulation is poorly understood, and further insight into Rgg function is anticipated to be of great importance for the understanding of both regulatory-network architecture and intercellular communication in Rgg-containing species. The results of this study on the Rgg2/3 QS circuit of S. pyogenes demonstrate that DNA binding of target promoters by the activator Rgg2 is directly inhibited by competitive binding by the repressor Rgg3, thereby preventing transcriptional activation of the target genes and premature induction of the QS circuit. This is a unique regulatory mechanism among Rgg proteins and other peptide-responsive QS regulators.
Figures


Similar articles
-
Structure-function studies of Rgg binding to pheromones and target promoters reveal a model of transcription factor interplay.Proc Natl Acad Sci U S A. 2020 Sep 29;117(39):24494-24502. doi: 10.1073/pnas.2008427117. Epub 2020 Sep 9. Proc Natl Acad Sci U S A. 2020. PMID: 32907945 Free PMC article.
-
Identification of Quorum-Sensing Inhibitors Disrupting Signaling between Rgg and Short Hydrophobic Peptides in Streptococci.mBio. 2015 May 12;6(3):e00393-15. doi: 10.1128/mBio.00393-15. mBio. 2015. PMID: 25968646 Free PMC article.
-
Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development.PLoS Pathog. 2011 Aug;7(8):e1002190. doi: 10.1371/journal.ppat.1002190. Epub 2011 Aug 4. PLoS Pathog. 2011. PMID: 21829369 Free PMC article.
-
Quorum sensing in group A Streptococcus.Front Cell Infect Microbiol. 2014 Sep 12;4:127. doi: 10.3389/fcimb.2014.00127. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25309879 Free PMC article. Review.
-
Peptide pheromone signaling in Streptococcus and Enterococcus.FEMS Microbiol Rev. 2014 May;38(3):473-92. doi: 10.1111/1574-6976.12046. Epub 2013 Oct 31. FEMS Microbiol Rev. 2014. PMID: 24118108 Free PMC article. Review.
Cited by
-
RstA Is a Major Regulator of Clostridioides difficile Toxin Production and Motility.mBio. 2019 Mar 12;10(2):e01991-18. doi: 10.1128/mBio.01991-18. mBio. 2019. PMID: 30862746 Free PMC article.
-
A Quorum Sensing-Regulated Protein Binds Cell Wall Components and Enhances Lysozyme Resistance in Streptococcus pyogenes.J Bacteriol. 2018 May 9;200(11):e00701-17. doi: 10.1128/JB.00701-17. Print 2018 Jun 1. J Bacteriol. 2018. PMID: 29555699 Free PMC article.
-
A novel chemical inducer of Streptococcus quorum sensing acts by inhibiting the pheromone-degrading endopeptidase PepO.J Biol Chem. 2018 Jan 19;293(3):931-940. doi: 10.1074/jbc.M117.810994. Epub 2017 Dec 4. J Biol Chem. 2018. PMID: 29203527 Free PMC article.
-
Nitric Oxide restricts iron availability and induces quorum sensing in Streptococcus pyogenes.Redox Biol. 2025 Jun 4;85:103699. doi: 10.1016/j.redox.2025.103699. Online ahead of print. Redox Biol. 2025. PMID: 40483894 Free PMC article.
-
The Proteomic and Transcriptomic Landscapes Altered by Rgg2/3 Activity in Streptococcus pyogenes.J Bacteriol. 2022 Nov 15;204(11):e0017522. doi: 10.1128/jb.00175-22. Epub 2022 Oct 31. J Bacteriol. 2022. PMID: 36314832 Free PMC article.
References
-
- Podbielski A, Kreikemeyer B. 2004. Cell density—dependent regulation: basic principles and effects on the virulence of Gram-positive cocci. Int. J. Infect. Dis. 8:81–95 - PubMed
-
- Rocha-Estrada J, Aceves-Diez AE, Guarneros G, de la Torre M. 2010. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 87:913–923 - PubMed
-
- Core L, Perego M. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509–1522 - PubMed
-
- D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655–662 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

