Rab7 mutants associated with Charcot-Marie-Tooth disease cause delayed growth factor receptor transport and altered endosomal and nuclear signaling
- PMID: 23188822
- PMCID: PMC3542998
- DOI: 10.1074/jbc.M112.417766
Rab7 mutants associated with Charcot-Marie-Tooth disease cause delayed growth factor receptor transport and altered endosomal and nuclear signaling
Abstract
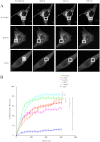
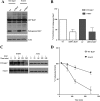
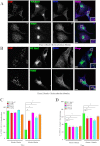
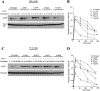
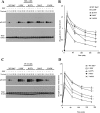
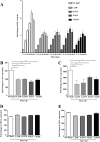
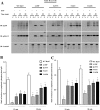
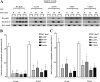
Rab7 belongs to the Ras superfamily of small GTPases and is a master regulator of early to late endocytic membrane transport. Four missense mutations in the late endosomal Rab7 GTPase (L129F, K157N, N161T, and V162M) cause the autosomal dominant peripheral neuropathy Charcot-Marie-Tooth type 2B (CMT2B) disease. As yet, the pathological mechanisms connecting mutant Rab7 protein expression to altered neuronal function are undefined. Here, we analyze the effects of Rab7 CMT2B mutants on epidermal growth factor (EGF)-dependent intracellular signaling and trafficking. Three different cell lines expressing Rab7 CMT2B mutants and stimulated with EGF exhibited delayed trafficking of EGF to LAMP1-positive late endosomes and lysosomes and slowed EGF receptor (EGFR) degradation. Expression of all Rab7 CMT2B mutants altered the Rab7 activation cycle, leading to enhanced and prolonged EGFR signaling as well as variable increases in p38 and ERK1/2 activation. However, due to reduced nuclear translocation of p38 and ERK1/2, the downstream nuclear activation of Elk-1 was decreased along with the expression of immediate early genes like c-fos and Egr-1 by the disease mutants. In conclusion, our results demonstrate that Rab7 CMT2B mutants impair growth factor receptor trafficking and, in turn, alter p38 and ERK1/2 signaling from perinuclear, clustered signaling endosomes. The resulting down-regulation of EGFR-dependent nuclear transcription that is crucial for normal axon outgrowth and peripheral innervation offers a crucial new mechanistic insight into disease pathogenesis that is relevant to other neurodegenerative diseases.
Figures

References
-
- Barisic N., Claeys K. G., Sirotković-Skerlev M., Löfgren A., Nelis E., De Jonghe P., Timmerman V. (2008) Charcot-Marie-Tooth disease. A clinico-genetic confrontation. Ann. Hum. Genet. 72, 416–441 - PubMed
-
- Shy M. E., Garbern J. Y., Kamholz J. (2002) Hereditary motor and sensory neuropathies. A biological perspective. Lancet Neurol. 1, 110–118 - PubMed
-
- Bienfait H. M., Baas F., Koelman J. H., de Haan R. J., van Engelen B. G., Gabreëls-Festen A. A., Ongerboer de Visser B. W., Meggouh F., Weterman M. A., De Jonghe P., Timmerman V., de Visser M. (2007) Phenotype of Charcot-Marie-Tooth disease Type 2. Neurology. 68, 1658–1667 - PubMed
-
- Snider M. D. (2003) A role for rab7 GTPase in growth factor-regulated cell nutrition and apoptosis. Mol. Cell. 12, 796–797 - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

