Functional analysis of Rad14p, a DNA damage recognition factor in nucleotide excision repair, in regulation of transcription in vivo
- PMID: 23188830
- PMCID: PMC3543029
- DOI: 10.1074/jbc.M112.413716
Functional analysis of Rad14p, a DNA damage recognition factor in nucleotide excision repair, in regulation of transcription in vivo
Abstract
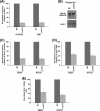
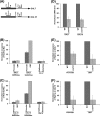
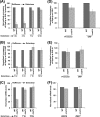
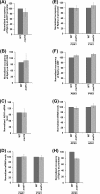
Rad14p is a DNA damage recognition factor in nucleotide excision repair. Intriguingly, we show here that Rad14p associates with the promoter of a galactose-inducible GAL1 gene after transcriptional induction in the absence of DNA lesion. Such an association of Rad14p facilitates the recruitment of TBP, TFIIH, and RNA polymerase II to the GAL1 promoter. Furthermore, the association of RNA polymerase II with the GAL1 promoter is significantly decreased in the absence of Rad14p, when the coding sequence was deleted. These results support the role of Rad14p in transcriptional initiation. Consistently, the level of GAL1 mRNA is significantly decreased in the absence of Rad14p. Similar results are also obtained at other galactose-inducible GAL genes such as GAL7 and GAL10. Likewise, Rad14p promotes transcription of other non-GAL genes such as CUP1, CTT1, and STL1 after transcriptional induction. However, the effect of Rad14p on the steady-state levels of transcription of GAL genes or constitutively active genes such as ADH1, PGK1, PYK1, and RPS5 is not observed. Thus, Rad14p promotes initial transcription but does not appear to regulate the steady-state level. Collectively, our results unveil a new role of Rad14p in stimulating transcription in addition to its well-known function in nucleotide excision repair.
Figures




Similar articles
-
Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response.Nucleic Acids Res. 2014 Sep;42(15):9892-907. doi: 10.1093/nar/gku703. Epub 2014 Aug 11. Nucleic Acids Res. 2014. PMID: 25114048 Free PMC article.
-
Epigenetic Transcriptional Memory of GAL Genes Depends on Growth in Glucose and the Tup1 Transcription Factor in Saccharomyces cerevisiae.Genetics. 2017 Aug;206(4):1895-1907. doi: 10.1534/genetics.117.201632. Epub 2017 Jun 12. Genetics. 2017. PMID: 28607146 Free PMC article.
-
Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes.J Biol Chem. 2004 Apr 2;279(14):14418-26. doi: 10.1074/jbc.M312004200. Epub 2004 Jan 19. J Biol Chem. 2004. PMID: 14734564 Free PMC article.
-
Histone acetylation, chromatin remodelling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae.Cell Cycle. 2005 Aug;4(8):1043-5. doi: 10.4161/cc.4.8.1928. Epub 2005 Aug 21. Cell Cycle. 2005. PMID: 16082210 Review.
-
Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: The ambiguous role of Rad26.DNA Repair (Amst). 2015 Dec;36:43-48. doi: 10.1016/j.dnarep.2015.09.006. Epub 2015 Sep 10. DNA Repair (Amst). 2015. PMID: 26429063 Review.
Cited by
-
Regulation of active genome integrity and expression by Rad26p.Nucleus. 2014;5(6):520-6. doi: 10.4161/nucl.36230. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484185 Free PMC article. Review.
-
Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response.Nucleic Acids Res. 2014 Sep;42(15):9892-907. doi: 10.1093/nar/gku703. Epub 2014 Aug 11. Nucleic Acids Res. 2014. PMID: 25114048 Free PMC article.
-
Comprehensive Synthetic Genetic Array Analysis of Alleles That Interact with Mutation of the Saccharomyces cerevisiae RecQ Helicases Hrq1 and Sgs1.G3 (Bethesda). 2020 Dec 3;10(12):4359-4368. doi: 10.1534/g3.120.401709. G3 (Bethesda). 2020. PMID: 33115720 Free PMC article.
-
Mechanisms of Antisense Transcription Initiation with Implications in Gene Expression, Genomic Integrity and Disease Pathogenesis.Noncoding RNA. 2019 Jan 21;5(1):11. doi: 10.3390/ncrna5010011. Noncoding RNA. 2019. PMID: 30669611 Free PMC article. Review.
-
Beyond the Trinity of ATM, ATR, and DNA-PK: Multiple Kinases Shape the DNA Damage Response in Concert With RNA Metabolism.Front Mol Biosci. 2019 Aug 2;6:61. doi: 10.3389/fmolb.2019.00061. eCollection 2019. Front Mol Biosci. 2019. PMID: 31428617 Free PMC article. Review.
References
-
- Prakash S., Prakash L. (2000) Nucleotide excision repair in yeast. Mutat. Res. 451, 13–24 - PubMed
-
- Guzder S. N., Sung P., Prakash L., Prakash S. (1996) Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J. Biol. Chem. 271, 8903–8910 - PubMed
-
- de Laat W. L., Jaspers N. G., Hoeijmakers J. H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 - PubMed
-
- Vermeulen W. (2011) Dynamics of mammalian NER proteins. DNA Repair 10, 760–771 - PubMed
-
- Bergoglio V., Magnaldo T. (2006) Nucleotide excision repair and related human diseases. Genome Dyn. 1, 35–52 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

