Genomic variation and its impact on gene expression in Drosophila melanogaster
- PMID: 23189034
- PMCID: PMC3499359
- DOI: 10.1371/journal.pgen.1003055
Genomic variation and its impact on gene expression in Drosophila melanogaster
Abstract
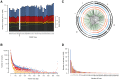
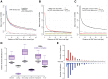
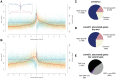
Understanding the relationship between genetic and phenotypic variation is one of the great outstanding challenges in biology. To meet this challenge, comprehensive genomic variation maps of human as well as of model organism populations are required. Here, we present a nucleotide resolution catalog of single-nucleotide, multi-nucleotide, and structural variants in 39 Drosophila melanogaster Genetic Reference Panel inbred lines. Using an integrative, local assembly-based approach for variant discovery, we identify more than 3.6 million distinct variants, among which were more than 800,000 unique insertions, deletions (indels), and complex variants (1 to 6,000 bp). While the SNP density is higher near other variants, we find that variants themselves are not mutagenic, nor are regions with high variant density particularly mutation-prone. Rather, our data suggest that the elevated SNP density around variants is mainly due to population-level processes. We also provide insights into the regulatory architecture of gene expression variation in adult flies by mapping cis-expression quantitative trait loci (cis-eQTLs) for more than 2,000 genes. Indels comprise around 10% of all cis-eQTLs and show larger effects than SNP cis-eQTLs. In addition, we identified two-fold more gene associations in males as compared to females and found that most cis-eQTLs are sex-specific, revealing a partial decoupling of the genomic architecture between the sexes as well as the importance of genetic factors in mediating sex-biased gene expression. Finally, we performed RNA-seq-based allelic expression imbalance analyses in the offspring of crosses between sequenced lines, which revealed that the majority of strong cis-eQTLs can be validated in heterozygous individuals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Mackay TFC, Stone EA, Ayroles JF (2009) The genetics of quantitative traits: challenges and prospects. Nat Rev Genet 10: 565–577. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

