DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing
- PMID: 23193179
- PMCID: PMC3561875
- DOI: 10.1101/gr.142067.112
DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing
Abstract
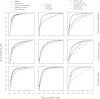
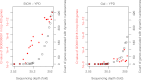
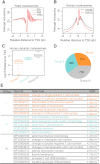
Recent developments in next-generation sequencing have enabled whole-genome profiling of nucleosome organizations. Although several algorithms for inferring nucleosome position from a single experimental condition have been available, it remains a challenge to accurately define dynamic nucleosomes associated with environmental changes. Here, we report a comprehensive bioinformatics pipeline, DANPOS, explicitly designed for dynamic nucleosome analysis at single-nucleotide resolution. Using both simulated and real nucleosome data, we demonstrated that bias correction in preliminary data processing and optimal statistical testing significantly enhances the functional interpretation of dynamic nucleosomes. The single-nucleotide resolution analysis of DANPOS allows us to detect all three categories of nucleosome dynamics, such as position shift, fuzziness change, and occupancy change, using a uniform statistical framework. Pathway analysis indicates that each category is involved in distinct biological functions. We also analyzed the influence of sequencing depth and suggest that even 200-fold coverage is probably not enough to identify all the dynamic nucleosomes. Finally, based on nucleosome data from the human hematopoietic stem cells (HSCs) and mouse embryonic stem cells (ESCs), we demonstrated that DANPOS is also robust in defining functional dynamic nucleosomes, not only in promoters, but also in distal regulatory regions in the mammalian genome.
Figures



References
-
- Bai FW, Anderson WA, Moo-Young M 2008. Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol Adv 26: 89–105 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
