Haustral fold segmentation with curvature-guided level set evolution
- PMID: 23193228
- PMCID: PMC3552127
- DOI: 10.1109/TBME.2012.2226242
Haustral fold segmentation with curvature-guided level set evolution
Abstract
Human colon has complex structures mostly because of the haustral folds. The folds are thin flat protrusions on the colon wall, which complicate the shape analysis for computer-aided detection (CAD) of colonic polyps. Fold segmentation may help reduce the structural complexity, and the folds can serve as an anatomic reference for computed tomographic colonography (CTC). Therefore, in this study, based on a model of the haustral fold boundaries, we developed a level-set approach to automatically segment the fold surfaces. To evaluate the developed fold segmentation algorithm, we first established the ground truth of haustral fold boundaries by experts' drawing on 15 patient CTC datasets without severe under/over colon distention from two medical centers. The segmentation algorithm successfully detected 92.7% of the folds in the ground truth. In addition to the sensitivity measure, we further developed a merit of segmented-area ratio (SAR), i.e., the ratio between the area of the intersection and union of the expert-drawn folds and the area of the automatically segmented folds, to measure the segmentation accuracy. The segmentation algorithm reached an average value of SAR = 86.2%, showing a good match with the ground truth on the fold surfaces. We believe the automatically segmented fold surfaces have the potential to benefit many postprocedures in CTC, such as CAD, taenia coli extraction, supine-prone registration, etc.
Figures
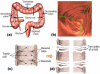

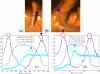
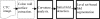
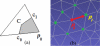
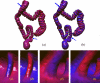

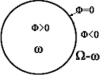
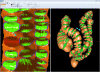
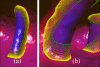


Similar articles
-
Haustral loop extraction for CT colonography using geodesics.Int J Comput Assist Radiol Surg. 2017 Mar;12(3):379-388. doi: 10.1007/s11548-016-1497-x. Epub 2016 Nov 16. Int J Comput Assist Radiol Surg. 2017. PMID: 27854032 Free PMC article.
-
Endoluminal surface registration for CT colonography using haustral fold matching.Med Image Anal. 2013 Dec;17(8):946-58. doi: 10.1016/j.media.2013.04.006. Epub 2013 Apr 27. Med Image Anal. 2013. PMID: 23845949 Free PMC article.
-
Automatic prone to supine haustral fold matching in CT colonography using a Markov random field model.Med Image Comput Comput Assist Interv. 2011;14(Pt 1):508-15. doi: 10.1007/978-3-642-23623-5_64. Med Image Comput Comput Assist Interv. 2011. PMID: 22003656
-
CAD techniques, challenges, and controversies in computed tomographic colonography.Abdom Imaging. 2005 Jan-Feb;30(1):26-41. doi: 10.1007/s00261-004-0244-x. Abdom Imaging. 2005. PMID: 15647868 Review.
-
Computer-aided detection for CT colonography: update 2007.Abdom Imaging. 2007 Sep-Oct;32(5):571-81. doi: 10.1007/s00261-007-9293-2. Abdom Imaging. 2007. PMID: 17690932 Review.
Cited by
-
A Novel Colon Wall Flattening Model for Computed Tomographic Colonography: Method and Validation.Comput Methods Biomech Biomed Eng Imaging Vis. 2014;13:1-14. doi: 10.1007/978-3-319-03590-1_1. Comput Methods Biomech Biomed Eng Imaging Vis. 2014. PMID: 25642397 Free PMC article.
-
Corresponding Supine and Prone Colon Visualization Using Eigenfunction Analysis and Fold Modeling.IEEE Trans Vis Comput Graph. 2017 Jan;23(1):751-760. doi: 10.1109/TVCG.2016.2598791. IEEE Trans Vis Comput Graph. 2017. PMID: 27875189 Free PMC article.
-
Haustral loop extraction for CT colonography using geodesics.Int J Comput Assist Radiol Surg. 2017 Mar;12(3):379-388. doi: 10.1007/s11548-016-1497-x. Epub 2016 Nov 16. Int J Comput Assist Radiol Surg. 2017. PMID: 27854032 Free PMC article.
References
-
- American Cancer Society . Cancer Facts & Figures 2008. American Cancer Society; Atlanta: 2008.
-
- Pickhardt P, Choi J, Hwang I, Butler J, Puckett M, Hildebrandt H, Wong R, Nugent P, Mysliwiec P, Schindler W. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. New England Journal of Medicine. 2003;349(23):2191–2200. - PubMed
-
- Taylor S, Iinuma G, Saito Y, Zhang J, Halligan S. CT colonography: computer-aided detection of morphologically flat T1 colonic carcinoma. European Radiology. 2008;18(8):1666–1673. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

