NCBI Epigenomics: what's new for 2013
- PMID: 23193265
- PMCID: PMC3531100
- DOI: 10.1093/nar/gks1171
NCBI Epigenomics: what's new for 2013
Abstract
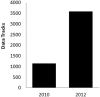
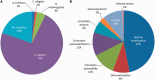
The Epigenomics resource at the National Center for Biotechnology Information (NCBI) has been created to serve as a comprehensive public repository for whole-genome epigenetic data sets (www.ncbi.nlm.nih.gov/epigenomics). We have constructed this resource by selecting the subset of epigenetics-specific data from the Gene Expression Omnibus (GEO) database and then subjecting them to further review and annotation. Associated data tracks can be viewed using popular genome browsers or downloaded for local analysis. We have performed extensive user testing throughout the development of this resource, and new features and improvements are continuously being implemented based on the results. We have made substantial usability improvements to user interfaces, enhanced functionality, made identification of data tracks of interest easier and created new tools for preliminary data analyses. Additionally, we have made efforts to enhance the integration between the Epigenomics resource and other NCBI databases, including the Gene database and PubMed. Data holdings have also increased dramatically since the initial publication describing the NCBI Epigenomics resource and currently consist of >3700 viewable and downloadable data tracks from 955 biological sources encompassing five well-studied species. This updated manuscript highlights these changes and improvements.
Figures
Similar articles
-
NCBI Epigenomics: a new public resource for exploring epigenomic data sets.Nucleic Acids Res. 2011 Jan;39(Database issue):D908-12. doi: 10.1093/nar/gkq1146. Epub 2010 Nov 12. Nucleic Acids Res. 2011. PMID: 21075792 Free PMC article.
-
NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update.Nucleic Acids Res. 2024 Jan 5;52(D1):D138-D144. doi: 10.1093/nar/gkad965. Nucleic Acids Res. 2024. PMID: 37933855 Free PMC article.
-
Database resources of the National Center for Biotechnology Information.Nucleic Acids Res. 2014 Jan;42(Database issue):D7-17. doi: 10.1093/nar/gkt1146. Epub 2013 Nov 19. Nucleic Acids Res. 2014. PMID: 24259429 Free PMC article.
-
NCBI Taxonomy: a comprehensive update on curation, resources and tools.Database (Oxford). 2020 Jan 1;2020:baaa062. doi: 10.1093/database/baaa062. Database (Oxford). 2020. PMID: 32761142 Free PMC article. Review.
-
Gene expression omnibus: microarray data storage, submission, retrieval, and analysis.Methods Enzymol. 2006;411:352-69. doi: 10.1016/S0076-6879(06)11019-8. Methods Enzymol. 2006. PMID: 16939800 Free PMC article. Review.
Cited by
-
DNApod: DNA polymorphism annotation database from next-generation sequence read archives.PLoS One. 2017 Feb 24;12(2):e0172269. doi: 10.1371/journal.pone.0172269. eCollection 2017. PLoS One. 2017. PMID: 28234924 Free PMC article.
-
A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics.PLoS One. 2013 Dec 6;8(12):e81148. doi: 10.1371/journal.pone.0081148. eCollection 2013. PLoS One. 2013. PMID: 24324667 Free PMC article.
-
Omics-based molecular techniques in oral pathology centred cancer: prospect and challenges in Africa.Cancer Cell Int. 2017 Jun 5;17:61. doi: 10.1186/s12935-017-0432-8. eCollection 2017. Cancer Cell Int. 2017. PMID: 28592923 Free PMC article. Review.
-
Epigenetic targets in B- and T-cell lymphomas: latest developments.Ther Adv Hematol. 2023 May 30;14:20406207231173485. doi: 10.1177/20406207231173485. eCollection 2023. Ther Adv Hematol. 2023. PMID: 37273421 Free PMC article. Review.
-
EpiFactors: a comprehensive database of human epigenetic factors and complexes.Database (Oxford). 2015 Jul 7;2015:bav067. doi: 10.1093/database/bav067. Print 2015. Database (Oxford). 2015. PMID: 26153137 Free PMC article.
References
-
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20.
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Ng RK, Gurdon JB. Epigenetic inheritance of cell differentiation status. Cell Cycle. 2008;7:1173–1177. - PubMed
-
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 2009;10:192–206. - PubMed

