DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications
- PMID: 23193290
- PMCID: PMC3531199
- DOI: 10.1093/nar/gks1229
DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications
Abstract
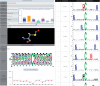
Protein modification is an extremely important post-translational regulation that adjusts the physical and chemical properties, conformation, stability and activity of a protein; thus altering protein function. Due to the high throughput of mass spectrometry (MS)-based methods in identifying site-specific post-translational modifications (PTMs), dbPTM (http://dbPTM.mbc.nctu.edu.tw/) is updated to integrate experimental PTMs obtained from public resources as well as manually curated MS/MS peptides associated with PTMs from research articles. Version 3.0 of dbPTM aims to be an informative resource for investigating the substrate specificity of PTM sites and functional association of PTMs between substrates and their interacting proteins. In order to investigate the substrate specificity for modification sites, a newly developed statistical method has been applied to identify the significant substrate motifs for each type of PTMs containing sufficient experimental data. According to the data statistics in dbPTM, >60% of PTM sites are located in the functional domains of proteins. It is known that most PTMs can create binding sites for specific protein-interaction domains that work together for cellular function. Thus, this update integrates protein-protein interaction and domain-domain interaction to determine the functional association of PTM sites located in protein-interacting domains. Additionally, the information of structural topologies on transmembrane (TM) proteins is integrated in dbPTM in order to delineate the structural correlation between the reported PTM sites and TM topologies. To facilitate the investigation of PTMs on TM proteins, the PTM substrate sites and the structural topology are graphically represented. Also, literature information related to PTMs, orthologous conservations and substrate motifs of PTMs are also provided in the resource. Finally, this version features an improved web interface to facilitate convenient access to the resource.
Figures
References
-
- Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003;21:255–261. - PubMed
-
- Farriol-Mathis N, Garavelli JS, Boeckmann B, Duvaud S, Gasteiger E, Gateau A, Veuthey AL, Bairoch A. Annotation of post-translational modifications in the Swiss-Prot knowledge base. Proteomics. 2004;4:1537–1550. - PubMed
-
- Seo J, Lee KJ. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004;37:35–44. - PubMed
-
- Wurgler-Murphy SM, King DM, Kennelly PJ. The Phosphorylation Site Database: a guide to the serine-, threonine-, and/or tyrosine-phosphorylated proteins in prokaryotic organisms. Proteomics. 2004;4:1562–1570. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous