DbVar and DGVa: public archives for genomic structural variation
- PMID: 23193291
- PMCID: PMC3531204
- DOI: 10.1093/nar/gks1213
DbVar and DGVa: public archives for genomic structural variation
Abstract
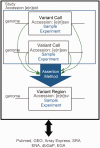
Much has changed in the last two years at DGVa (http://www.ebi.ac.uk/dgva) and dbVar (http://www.ncbi.nlm.nih.gov/dbvar). We are now processing direct submissions rather than only curating data from the literature and our joint study catalog includes data from over 100 studies in 11 organisms. Studies from human dominate with data from control and case populations, tumor samples as well as three large curated studies derived from multiple sources. During the processing of these data, we have made improvements to our data model, submission process and data representation. Additionally, we have made significant improvements in providing access to these data via web and FTP interfaces.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

