MicroRNA Response Elements-Mediated miRNA-miRNA Interactions in Prostate Cancer
- PMID: 23193399
- PMCID: PMC3502784
- DOI: 10.1155/2012/839837
MicroRNA Response Elements-Mediated miRNA-miRNA Interactions in Prostate Cancer
Abstract
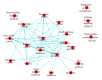
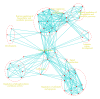
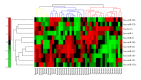
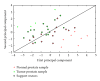
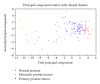
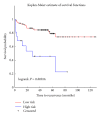
The cell is a highly organized system of interacting molecules including proteins, mRNAs, and miRNAs. Analyzing the cell from a systems perspective by integrating different types of data helps revealing the complexity of diseases. Although there is emerging evidence that microRNAs have a functional role in cancer, the role of microRNAs in mediating cancer progression and metastasis remains not fully explored. As the amount of available miRNA and mRNA gene expression data grows, more systematic methods combining gene expression and biological networks become necessary to explore miRNA function. In this work I integrated functional miRNA-target interactions with mRNA and miRNA expression to infer mRNA-mediated miRNA-miRNA interactions. The inferred network represents miRNA modulation through common targets. The network is used to characterize the functional role of microRNA response element (MRE) to mediate interactions between miRNAs targeting the MRE. Results revealed that miRNA-1 is a key player in regulating prostate cancer progression. 11 miRNAs were identified as diagnostic and prognostic biomarkers that act as tumor suppressor miRNAs. This work demonstrates the utility of a network analysis as opposed to differential expression to find important miRNAs that regulate prostate cancer.
Figures



References
-
- Ruvkun G. Molecular biology: glimpses of a tiny RNA world. Science. 2001;294(5543):797–799. - PubMed
-
- Sevli S, Uzumcu A, Solak M, Ittmann M, Ozen M. The function of microRNAs, small but potent molecules, in human prostate cancer. Prostate Cancer and Prostatic Diseases. 2010;13(3):208–217. - PubMed
-
- Gordanpour A, Nam RK, Sugar L, Seth A. MicroRNAs in prostate cancer: from biomarkers to molecularly-based therapeutics. Prostate Cancer and Prostatic Diseases. 2012;1:p. 6. - PubMed
-
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Reviews Genetics. 2004;5(8):p. 631.
-
- Pang Y, Young CYF, Yuan H. MicroRNAs and prostate cancer. Acta Biochimica et Biophysica Sinica. 2010;42(6):363–369. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

