Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS)
- PMID: 23194976
- PMCID: PMC3613897
- DOI: 10.1016/j.jbior.2012.09.005
Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS)
Abstract
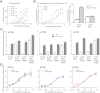
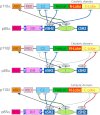
The catalytic subunits of all class IA phosphoinositide 3-kinases (PI3Ks) associate with identical p85-related subunits and phosphorylate PIP2 yielding PIP3, but they can vary greatly in the signaling pathways in which they participate. The binding of the p85 subunit to the p110 catalytic subunits is constitutive, and this inhibits activity, but some of the inhibitory contacts are reversible and subject to regulation. Interaction with phosphotyrosine-containing peptides (RTK-pY) releases a subset of these inhibitory contacts. Hydrogen/deuterium exchange mass spectrometry (HDX-MS) provides a map of the dynamic interactions unique to each of the isotypes. RTK-pY binding exposes the p110 helical domains for all class IA enzymes (due to release of the nSH2 contact) and exposes the C-lobe of the kinase domains of p110β and p110δ (resulting from release of the cSH2 contact). Consistent with this, our in vitro assays show that all class IA isoforms are inhibited by the nSH2, but only p110β and p110δ are inhibited by the cSH2. While a C2/iSH2 inhibitory contact exists in all isoforms, HDX indicates that p110β releases this contact most readily. The unique dynamic relationships of the different p110 isozymes to the p85 subunit may facilitate new strategies for specific inhibitors of the PI3Ks.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

