An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants
- PMID: 23197803
- PMCID: PMC3561014
- DOI: 10.1104/pp.112.209254
An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants
Abstract
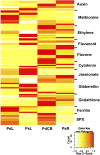
Phosphorus, in its orthophosphate form (P(i)), is one of the most limiting macronutrients in soils for plant growth and development. However, the whole-genome molecular mechanisms contributing to plant acclimation to P(i) deficiency remain largely unknown. White lupin (Lupinus albus) has evolved unique adaptations for growth in P(i)-deficient soils, including the development of cluster roots to increase root surface area. In this study, we utilized RNA-Seq technology to assess global gene expression in white lupin cluster roots, normal roots, and leaves in response to P(i) supply. We de novo assembled 277,224,180 Illumina reads from 12 complementary DNA libraries to build what is to our knowledge the first white lupin gene index (LAGI 1.0). This index contains 125,821 unique sequences with an average length of 1,155 bp. Of these sequences, 50,734 were transcriptionally active (reads per kilobase per million reads ≥ 3), representing approximately 7.8% of the white lupin genome, using the predicted genome size of Lupinus angustifolius as a reference. We identified a total of 2,128 sequences differentially expressed in response to P(i) deficiency with a 2-fold or greater change and P ≤ 0.05. Twelve sequences were consistently differentially expressed due to P(i) deficiency stress in three species, Arabidopsis (Arabidopsis thaliana), potato (Solanum tuberosum), and white lupin, making them ideal candidates to monitor the P(i) status of plants. Additionally, classic physiological experiments were coupled with RNA-Seq data to examine the role of cytokinin and gibberellic acid in P(i) deficiency-induced cluster root development. This global gene expression analysis provides new insights into the biochemical and molecular mechanisms involved in the acclimation to P(i) deficiency.
Figures





References
-
- Amador V, Monte E, García-Martínez J-L, Prat S. (2001) Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106: 343–354 - PubMed
-
- Andersson MX, Stridh MH, Larsson KE, Liljenberg C, Sandelius AS. (2003) Phosphate-deficient oat replaces a major portion of the plasma membrane phospholipids with the galactolipid digalactosyldiacylglycerol. FEBS Lett 537: 128–132 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

