Noninvasive Fetal Trisomy (NIFTY) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies
- PMID: 23198897
- PMCID: PMC3544640
- DOI: 10.1186/1755-8794-5-57
Noninvasive Fetal Trisomy (NIFTY) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies
Abstract
Background: Conventional prenatal screening tests, such as maternal serum tests and ultrasound scan, have limited resolution and accuracy.
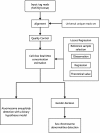
Methods: We developed an advanced noninvasive prenatal diagnosis method based on massively parallel sequencing. The Noninvasive Fetal Trisomy (NIFTY) test, combines an optimized Student's t-test with a locally weighted polynomial regression and binary hypotheses. We applied the NIFTY test to 903 pregnancies and compared the diagnostic results with those of full karyotyping.
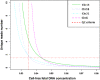
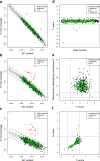
Results: 16 of 16 trisomy 21, 12 of 12 trisomy 18, two of two trisomy 13, three of four 45, X, one of one XYY and two of two XXY abnormalities were correctly identified. But one false positive case of trisomy 18 and one false negative case of 45, X were observed. The test performed with 100% sensitivity and 99.9% specificity for autosomal aneuploidies and 85.7% sensitivity and 99.9% specificity for sex chromosomal aneuploidies. Compared with three previously reported z-score approaches with/without GC-bias removal and with internal control, the NIFTY test was more accurate and robust for the detection of both autosomal and sex chromosomal aneuploidies in fetuses.
Conclusion: Our study demonstrates a powerful and reliable methodology for noninvasive prenatal diagnosis.
Figures

References
-
- Bouchlariotou S, Tsikouras P, Dimitraki M, Athanasiadis A, Papoulidis I, Maroulis G, Liberis A, Liberis V. Turner’s syndrome and pregnancy: has the 45, X/47, XXX mosaicism a different prognosis? Own clinical experience and literature review. J Matern Fetal Neonatal Med. 2011;24(5):668–672. doi: 10.3109/14767058.2010.520769. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

