Pharmacogenomics of chemotherapeutic susceptibility and toxicity
- PMID: 23199206
- PMCID: PMC3580423
- DOI: 10.1186/gm391
Pharmacogenomics of chemotherapeutic susceptibility and toxicity
Abstract
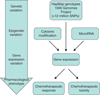
The goal of personalized medicine is to tailor a patient's treatment strategy on the basis of his or her unique genetic make-up. The field of oncology is beginning to incorporate many of the strategies of personalized medicine, especially within the realm of pharmacogenomics, which is the study of how inter-individual genetic variation determines drug response or toxicity. A main objective of pharmacogenomics is to facilitate physician decision-making regarding optimal drug selection, dose and treatment duration on a patient-by-patient basis. Recent advances in genome-wide genotyping and sequencing technologies have supported the discoveries of a number of pharmacogenetic markers that predict response to chemotherapy. However, effectively implementing these pharmacogenetic markers in the clinic remains a major challenge. This review focuses on the contribution of germline genetic variation to chemotherapeutic toxicity and response, and discusses the utility of genome-wide association studies and use of lymphoblastoid cell lines (LCLs) in pharmacogenomic studies. Furthermore, we highlight several recent examples of genetic variants associated with chemotherapeutic toxicity or response in both patient cohorts and LCLs, and discuss the challenges and future directions of pharmacogenomic discovery for cancer treatment.
Keywords: International HapMap Project; Pharmacogenomics; chemotherapeutics; clinical translation; genome-wide association studies.
Figures

References
-
- Ramsey LB, Bruun GH, Yang W, Treviño LR, Vattathil S, Scheet P, Cheng C, Rosner GL, Giacomini KM, Fan Y, Sparreboom A, Mikkelsen TS, Corydon TJ, Pui CH, Evans WE, Relling MV. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. - DOI - PMC - PubMed
-
- Innocenti F, Owzar K, Cox NL, Evans P, Kubo M, Zembutsu H, Jiang C, Hollis D, Mushiroda T, Li L, Friedman P, Wang L, Glubb D, Hiacomini KM, McLeod HL, Goldberg RM, Schilsky RL, Kindler HL Makamura Y, Ratain MJ. A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303. Clin Cancer Res. 2012;18:577–584. doi: 10.1158/1078-0432.CCR-11-1387. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

