The human crystallin gene families
- PMID: 23199295
- PMCID: PMC3554465
- DOI: 10.1186/1479-7364-6-26
The human crystallin gene families
Abstract
Crystallins are the abundant, long-lived proteins of the eye lens. The major human crystallins belong to two different superfamilies: the small heat-shock proteins (α-crystallins) and the βγ-crystallins. During evolution, other proteins have sometimes been recruited as crystallins to modify the properties of the lens. In the developing human lens, the enzyme betaine-homocysteine methyltransferase serves such a role. Evolutionary modification has also resulted in loss of expression of some human crystallin genes or of specific splice forms. Crystallin organization is essential for lens transparency and mutations; even minor changes to surface residues can cause cataract and loss of vision.
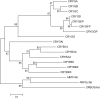
Figures


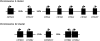
Similar articles
-
sHSP in the eye lens: crystallin mutations, cataract and proteostasis.Int J Biochem Cell Biol. 2012 Oct;44(10):1687-97. doi: 10.1016/j.biocel.2012.02.015. Epub 2012 Mar 2. Int J Biochem Cell Biol. 2012. PMID: 22405853 Review.
-
Differential role of arginine mutations on the structure and functions of α-crystallin.Biochim Biophys Acta. 2016 Jan;1860(1 Pt B):199-210. doi: 10.1016/j.bbagen.2015.06.004. Epub 2015 Jun 14. Biochim Biophys Acta. 2016. PMID: 26080000 Free PMC article. Review.
-
Probing the changes in gene expression due to α-crystallin mutations in mouse models of hereditary human cataract.PLoS One. 2018 Jan 16;13(1):e0190817. doi: 10.1371/journal.pone.0190817. eCollection 2018. PLoS One. 2018. PMID: 29338044 Free PMC article.
-
Effects of alpha-crystallin on lens cell function and cataract pathology.Curr Mol Med. 2009 Sep;9(7):887-92. doi: 10.2174/156652409789105598. Curr Mol Med. 2009. PMID: 19860667 Review.
-
Evolutionary Origins of Pax6 Control of Crystallin Genes.Genome Biol Evol. 2017 Aug 1;9(8):2075-2092. doi: 10.1093/gbe/evx153. Genome Biol Evol. 2017. PMID: 28903537 Free PMC article.
Cited by
-
Evolution of crystallins for a role in the vertebrate eye lens.Protein Sci. 2013 Apr;22(4):367-80. doi: 10.1002/pro.2229. Epub 2013 Feb 26. Protein Sci. 2013. PMID: 23389822 Free PMC article. Review.
-
Divalent Cations and the Divergence of βγ-Crystallin Function.Biochemistry. 2019 Nov 12;58(45):4505-4518. doi: 10.1021/acs.biochem.9b00507. Epub 2019 Nov 1. Biochemistry. 2019. PMID: 31647219 Free PMC article.
-
Whole Exome Sequencing of 20 Spanish Families: Candidate Genes for Non-Syndromic Pediatric Cataracts.Int J Mol Sci. 2023 Jul 13;24(14):11429. doi: 10.3390/ijms241411429. Int J Mol Sci. 2023. PMID: 37511188 Free PMC article.
-
A genomic deletion encompassing CRYBB2-CRYBB2P1 is responsible for autosomal recessive congenital cataracts.Hum Genome Var. 2022 Sep 8;9(1):31. doi: 10.1038/s41439-022-00208-7. Hum Genome Var. 2022. PMID: 36075891 Free PMC article.
-
Enzyme complexity in intermediary metabolism.J Inherit Metab Dis. 2015 Jul;38(4):721-7. doi: 10.1007/s10545-015-9821-0. Epub 2015 Feb 21. J Inherit Metab Dis. 2015. PMID: 25700988 Review.
References
-
- Wistow G, Slingsby C. Structure and evolution of crystallins. The Encyclopedia of the Eye. 2010;2:229–238.
-
- Harding JJ, Crabbe MJC. The lens: development, proteins, metabolism and cataract. Eye. 1984;1B:207–492.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

