Evolution of Bcl-2 homology motifs: homology versus homoplasy
- PMID: 23199982
- PMCID: PMC3582728
- DOI: 10.1016/j.tcb.2012.10.010
Evolution of Bcl-2 homology motifs: homology versus homoplasy
Abstract
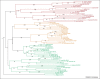
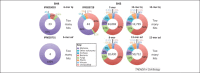
Bcl-2 family proteins regulate apoptosis in animals. This protein family includes several homologous proteins and a collection of other proteins lacking sequence similarity except for a Bcl-2 homology (BH)3 motif. Thus, membership in the Bcl-2 family requires only one of the four BH motifs. On this basis, a growing number of diverse BH3-only proteins are being reported. Although compelling cell biological and biophysical evidence validates many BH3-only proteins, claims of significant BH3 sequence similarity are often unfounded. Computational and phylogenetic analyses suggest that only some BH3 motifs arose by divergent evolution from a common ancestor (homology), whereas others arose by convergent evolution or random coincidence (homoplasy), challenging current assumptions about which proteins constitute the extended Bcl-2 family.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Hengartner M.O., Horvitz H.R. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell. 1994;76:665–676. - PubMed
-
- Tsujimoto Y. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. - PubMed
-
- Chittenden T. Induction of apoptosis by the Bcl-2 homologue Bak. Nature. 1995;374:733–736. - PubMed
-
- Oltvai Z.N. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

