GABA binding to an insect GABA receptor: a molecular dynamics and mutagenesis study
- PMID: 23200041
- PMCID: PMC3512037
- DOI: 10.1016/j.bpj.2012.10.016
GABA binding to an insect GABA receptor: a molecular dynamics and mutagenesis study
Abstract
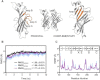
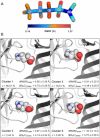
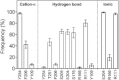
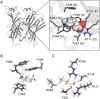
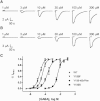
RDL receptors are GABA-activated inhibitory Cys-loop receptors found throughout the insect CNS. They are a key target for insecticides. Here, we characterize the GABA binding site in RDL receptors using computational and electrophysiological techniques. A homology model of the extracellular domain of RDL was generated and GABA docked into the binding site. Molecular dynamics simulations predicted critical GABA binding interactions with aromatic residues F206, Y254, and Y109 and hydrophilic residues E204, S176, R111, R166, S176, and T251. These residues were mutated, expressed in Xenopus oocytes, and their functions assessed using electrophysiology. The data support the binding mechanism provided by the simulations, which predict that GABA forms many interactions with binding site residues, the most significant of which are cation-π interactions with F206 and Y254, H-bonds with E204, S205, R111, S176, T251, and ionic interactions with R111 and E204. These findings clarify the roles of a range of residues in binding GABA in the RDL receptor, and also show that molecular dynamics simulations are a useful tool to identify specific interactions in Cys-loop receptors.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Thompson A.J., Lester H.A., Lummis S.C.R. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010;43:449–499. - PubMed
-
- Lummis S.C.R. Locating GABA in GABA receptor binding sites. Biochem. Soc. Trans. 2009;37:1343–1346. - PubMed
-
- Woodward R.M., Polenzani L., Miledi R. Characterization of bicuculline/baclofen-insensitive (ρ-like) γ-aminobutyric acid receptors expressed in Xenopus oocytes. II. Pharmacology of γ-aminobutyric acidA and γ-aminobutyric acidB receptor agonists and antagonists. Mol. Pharmacol. 1993;43:609–625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

