Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation
- PMID: 23200050
- PMCID: PMC3512040
- DOI: 10.1016/j.bpj.2012.10.013
Coupled flexibility change in cytochrome P450cam substrate binding determined by neutron scattering, NMR, and molecular dynamics simulation
Abstract
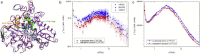
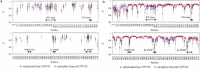
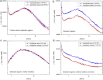
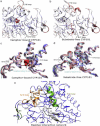
Neutron scattering and nuclear magnetic resonance relaxation experiments are combined with molecular dynamics (MD) simulations in a novel, to our knowledge, approach to investigate the change in internal dynamics on substrate (camphor) binding to a protein (cytochrome P450cam). The MD simulations agree well with both the neutron scattering, which furnishes information on global flexibility, and the nuclear magnetic resonance data, which provides residue-specific order parameters. Decreased fluctuations are seen in the camphor-bound form using all three techniques, dominated by changes in specific regions of the protein. The combined experimental and simulation results permit a detailed description of the dynamical change, which involves modifications in the coupling between the dominant regions and concomitant substrate access channel closing, via specific salt-bridge, hydrogen-bonding, and hydrophobic interactions. The work demonstrates how the combination of complementary experimental spectroscopies with MD simulation can provide an in-depth description of functional dynamical protein changes.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms.J Mol Biol. 2000 Nov 10;303(5):797-811. doi: 10.1006/jmbi.2000.4154. J Mol Biol. 2000. PMID: 11061976
-
Experimentally restrained molecular dynamics simulations for characterizing the open states of cytochrome P450cam.Biochemistry. 2011 Mar 15;50(10):1664-71. doi: 10.1021/bi101820d. Epub 2011 Feb 8. Biochemistry. 2011. PMID: 21265500 Free PMC article.
-
Mapping the Substrate Recognition Pathway in Cytochrome P450.J Am Chem Soc. 2018 Dec 19;140(50):17743-17752. doi: 10.1021/jacs.8b10840. Epub 2018 Dec 10. J Am Chem Soc. 2018. PMID: 30479124
-
Cytochrome P450cam: crystallography, oxygen activation, and electron transfer.FASEB J. 1992 Jan 6;6(2):674-9. doi: 10.1096/fasebj.6.2.1537455. FASEB J. 1992. PMID: 1537455 Review.
-
Dynamic processes in biological membrane mimics revealed by quasielastic neutron scattering.Chem Phys Lipids. 2017 Aug;206:28-42. doi: 10.1016/j.chemphyslip.2017.05.009. Epub 2017 Jun 1. Chem Phys Lipids. 2017. PMID: 28579420 Review.
Cited by
-
Unconstrained Enhanced Sampling for Free Energy Calculations of Biomolecules: A Review.Mol Simul. 2016;42(13):1046-1055. doi: 10.1080/08927022.2015.1121541. Epub 2016 Jul 5. Mol Simul. 2016. PMID: 27453631 Free PMC article.
-
Comparison of intrinsic dynamics of cytochrome p450 proteins using normal mode analysis.Protein Sci. 2015 Sep;24(9):1495-507. doi: 10.1002/pro.2737. Epub 2015 Jul 16. Protein Sci. 2015. PMID: 26130403 Free PMC article.
-
Zaccai neutron resilience and site-specific hydration dynamics in a globular protein.Eur Phys J E Soft Matter. 2013 Jul;36(7):72. doi: 10.1140/epje/i2013-13072-5. Epub 2013 Jul 16. Eur Phys J E Soft Matter. 2013. PMID: 23852576
-
Dynamics of CYP51: implications for function and inhibitor design.J Mol Recognit. 2015 Feb;28(2):59-73. doi: 10.1002/jmr.2412. Epub 2015 Jan 20. J Mol Recognit. 2015. PMID: 25601796 Free PMC article.
-
Acceleration of biomolecular kinetics in Gaussian accelerated molecular dynamics.J Chem Phys. 2018 Aug 21;149(7):072308. doi: 10.1063/1.5024217. J Chem Phys. 2018. PMID: 30134710 Free PMC article.
References
-
- Shaik S., Cohen S., Thiel W. P450 enzymes: their structure, reactivity, and selectivity modeled by QM/MM calculations. Chem. Rev. 2010;110:949–1017. - PubMed
-
- Schlichting I., Berendzen J., Sligar S.G. The catalytic pathway of cytochrome p450cam at atomic resolution. Science. 2000;287:1615–1622. - PubMed
-
- Poulos T.L., Finzel B.C., Howard A.J. Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry. 1986;25:5314–5322. - PubMed
-
- Poulos T.L., Finzel B.C., Howard A.J. High-resolution crystal structure of cytochrome P450cam. J. Mol. Biol. 1987;195:687–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

