Force-clamp analysis techniques give highest rank to stretched exponential unfolding kinetics in ubiquitin
- PMID: 23200055
- PMCID: PMC3512049
- DOI: 10.1016/j.bpj.2012.10.022
Force-clamp analysis techniques give highest rank to stretched exponential unfolding kinetics in ubiquitin
Abstract
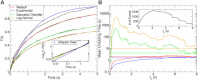
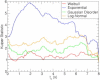
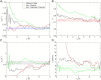
Force-clamp spectroscopy reveals the unfolding and disulfide bond rupture times of single protein molecules as a function of the stretching force, point mutations, and solvent conditions. The statistics of these times reveal whether the protein domains are independent of one another, the mechanical hierarchy in the polyprotein chain, and the functional form of the probability distribution from which they originate. It is therefore important to use robust statistical tests to decipher the correct theoretical model underlying the process. Here, we develop multiple techniques to compare the well-established experimental data set on ubiquitin with existing theoretical models as a case study. We show that robustness against filtering, agreement with a maximum likelihood function that takes into account experimental artifacts, the Kuiper statistic test, and alignment with synthetic data all identify the Weibull or stretched exponential distribution as the best fitting model. Our results are inconsistent with recently proposed models of Gaussian disorder in the energy landscape or noise in the applied force as explanations for the observed nonexponential kinetics. Because the physical model in the fit affects the characteristic unfolding time, these results have important implications on our understanding of the biological function of proteins.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Fernandez J.M., Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–1678. - PubMed
-
- Perez-Jimenez R., Garcia-Manyes S., Fernandez J.M. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J. Biol. Chem. 2006;281:40010–40014. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

