Taking a cellular road-trip: mRNA transport and anchoring
- PMID: 23200723
- PMCID: PMC3583371
- DOI: 10.1016/j.ceb.2012.08.015
Taking a cellular road-trip: mRNA transport and anchoring
Abstract
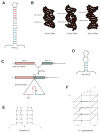
mRNA localization is a crucial mechanism for post-transcriptional control of gene expression used in numerous cellular contexts to generate asymmetric enrichment of an encoded protein. This process has emerged as a fundamental regulatory mechanism that operates in a wide range of organisms to control an array of cellular processes. Recently, significant advances have been made in our understanding of the mechanisms that regulate several steps in the mRNA localization pathway. Here we discuss the progress made in understanding localization element recognition, paying particular attention to the role of RNA structure. We also consider the function of mRNP granules in mRNA transport, as well as new results pointing to roles for the endocytic pathway in mRNA localization.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
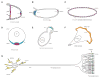
Similar articles
-
A cytoplasmic complex mediates specific mRNA recognition and localization in yeast.PLoS Biol. 2011 Apr;9(4):e1000611. doi: 10.1371/journal.pbio.1000611. Epub 2011 Apr 19. PLoS Biol. 2011. PMID: 21526221 Free PMC article.
-
Sequence-Independent Self-Assembly of Germ Granule mRNAs into Homotypic Clusters.Mol Cell. 2020 Jun 4;78(5):941-950.e12. doi: 10.1016/j.molcel.2020.05.008. Epub 2020 May 27. Mol Cell. 2020. PMID: 32464092 Free PMC article.
-
mRNA localization: an orchestration of assembly, traffic and synthesis.Traffic. 2013 Jan;14(1):2-14. doi: 10.1111/tra.12004. Epub 2012 Sep 13. Traffic. 2013. PMID: 22913533 Free PMC article. Review.
-
Principles and properties of eukaryotic mRNPs.Mol Cell. 2014 May 22;54(4):547-58. doi: 10.1016/j.molcel.2014.04.033. Mol Cell. 2014. PMID: 24856220 Review.
-
From birth to death: the complex lives of eukaryotic mRNAs.Science. 2005 Sep 2;309(5740):1514-8. doi: 10.1126/science.1111443. Science. 2005. PMID: 16141059 Review.
Cited by
-
The Xenopus Maternal-to-Zygotic Transition from the Perspective of the Germline.Curr Top Dev Biol. 2015;113:271-303. doi: 10.1016/bs.ctdb.2015.07.021. Epub 2015 Aug 21. Curr Top Dev Biol. 2015. PMID: 26358876 Free PMC article. Review.
-
Global mRNA polarization regulates translation efficiency in the intestinal epithelium.Science. 2017 Sep 22;357(6357):1299-1303. doi: 10.1126/science.aan2399. Epub 2017 Aug 10. Science. 2017. PMID: 28798045 Free PMC article.
-
mRNA localization in metazoans: A structural perspective.RNA Biol. 2017 Nov 2;14(11):1473-1484. doi: 10.1080/15476286.2017.1338231. Epub 2017 Jul 31. RNA Biol. 2017. PMID: 28640665 Free PMC article. Review.
-
The mRNA dynamics underpinning translational control mechanisms of Drosophila melanogaster oogenesis.Biochem Soc Trans. 2024 Oct 30;52(5):2087-2099. doi: 10.1042/BST20231293. Biochem Soc Trans. 2024. PMID: 39263986 Free PMC article. Review.
-
Mapping translation 'hot-spots' in live cells by tracking single molecules of mRNA and ribosomes.Elife. 2016 Jan 13;5:e10415. doi: 10.7554/eLife.10415. Elife. 2016. PMID: 26760529 Free PMC article.
References
-
- Jeffery WR, Tomlinson CR, Brodeur RD. Localization of actin messenger RNA during early ascidian development. Dev Biol. 1983;99:408–417. - PubMed
-
- Tomlinson CR, Bates WR, Jeffery WR. Development of a muscle actin specified by maternal and zygotic mRNA in ascidian embryos. Dev Biol. 1987;123:470–482. - PubMed
-
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. - PubMed
-
- Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

