Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity
- PMID: 23200854
- PMCID: PMC3628775
- DOI: 10.1016/j.celrep.2012.10.025
Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity
Abstract
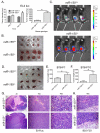
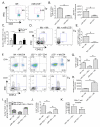
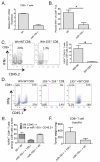
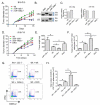
An increased understanding of antitumor immunity is necessary for improving cell-based immunotherapies against human cancers. Here, we investigated the roles of two immune system-expressed microRNAs (miRNAs), miR-155 and miR-146a, in the regulation of antitumor immune responses. Our results indicate that miR-155 promotes and miR-146a inhibits interferon γ (IFNγ) responses by T cells and reduces solid tumor growth in vivo. Using a double-knockout (DKO) mouse strain deficient in both miR-155 and miR-146a, we have also identified an epistatic relationship between these two miRNAs. DKO mice had defective T cell responses and tumor growth phenotypes similar to miR-155(-/-) mice. Further analysis of the T cell compartment revealed that miR-155 modulates IFNγ expression through a mechanism involving repression of Ship1. Our work reveals critical roles for miRNAs in the reciprocal regulation of CD4(+) and CD8(+) T cell-mediated antitumor immunity and demonstrates the dominant nature of miR-155 during its promotion of immune responses.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Cubillos-Ruiz JR, Baird JR, Tesone AJ, Rutkowski MR, Scarlett UK, Camposeco-Jacobs AL, Anadon-Arnillas J, Harwood NM, Korc M, Fiering SN, et al. Reprogramming tumor-associated dendritic cells in vivo using miRNA mimetics triggers protective immunity against ovarian cancer. Cancer Res. 2012;72:1683–1693. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

