High level of chromosomal aberration in ovarian cancer genome correlates with poor clinical outcome
- PMID: 23200914
- PMCID: PMC4364416
- DOI: 10.1016/j.ygyno.2012.11.031
High level of chromosomal aberration in ovarian cancer genome correlates with poor clinical outcome
Abstract
Objectives: Structural aberration in chromosomes characterizes almost all human solid cancers and analysis of those alterations may reveal the history of chromosomal instability. However, the clinical significance of massive chromosomal abnormality in ovarian high-grade serous carcinoma (HGSC) remains elusive. In this study, we addressed this issue by analyzing the genomic profiles in 455 ovarian HGSCs available from The Cancer Genome Atlas (TCGA).
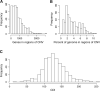
Methods: DNA copy number, mRNA expression, and clinical information were downloaded from the TCGA data portal. A chromosomal disruption index (CDI) was developed to summarize the extent of copy number aberrations across the entire genome. A Cox regression model was applied to identify factors associated with poor prognosis. Genes whose expression was associated with CDI were identified by a 2-stage multivariate linear regression and were used to find enriched pathways by Ingenuity Pathway Analysis.
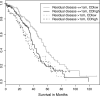
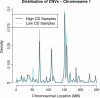
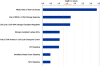
Results: Multivariate survival analysis showed that a higher CDI was significantly associated with a worse overall survival in patients. Interestingly, the pattern of DNA copy number alterations across all the chromosomes was similar between tumors with high and low CDI, suggesting they did not arise from different mechanisms. We also observed that expression of several genes was highly correlated with the CDI, even after adjusting for local copy number variation. We found that molecular pathways involving DNA damage response and mitosis were significantly enriched in these CDI-correlated genes.
Conclusion: Our results provide a new insight into the role of chromosomal rearrangement in the development of HGSC and the promise of applying CDI in risk-stratifying HGSC patients, perhaps for different clinical managements. The genes whose expression is correlated with CDI are worthy of further study to elucidate the mechanism of chromosomal instability in HGSC.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
References
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–76. - PubMed
-
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462(2–3):247–53. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

