Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome
- PMID: 23201123
- PMCID: PMC3704155
- DOI: 10.1016/j.molcel.2012.10.024
Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome
Abstract
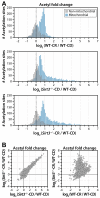
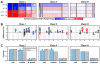
Calorie restriction (CR) extends life span in diverse species. Mitochondria play a key role in CR adaptation; however, the molecular details remain elusive. We developed and applied a quantitative mass spectrometry method to probe the liver mitochondrial acetyl-proteome during CR versus control diet in mice that were wild-type or lacked the protein deacetylase SIRT3. Quantification of 3,285 acetylation sites-2,193 from mitochondrial proteins-rendered a comprehensive atlas of the acetyl-proteome and enabled global site-specific, relative acetyl occupancy measurements between all four experimental conditions. Bioinformatic and biochemical analyses provided additional support for the effects of specific acetylation on mitochondrial protein function. Our results (1) reveal widespread reprogramming of mitochondrial protein acetylation in response to CR and SIRT3, (2) identify three biochemically distinct classes of acetylation sites, and (3) provide evidence that SIRT3 is a prominent regulator in CR adaptation by coordinately deacetylating proteins involved in diverse pathways of metabolism and mitochondrial maintenance.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Bathe OF, Shaykhutdinov R, Kopciuk K, Weljie AM, McKay A, Sutherland FR, Dixon E, Dunse N, Sotiropoulos D, Vogel HJ. Feasibility of identifying pancreatic cancer based on serum metabolomics. Cancer Epidemiol Biomarkers Prev. 2011;20:140–147. - PubMed
-
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012 - PubMed
-
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM080148/GM/NIGMS NIH HHS/United States
- R01 AG038679/AG/NIA NIH HHS/United States
- P30 CA014520/CA/NCI NIH HHS/United States
- R01 DK098672/DK/NIDDK NIH HHS/United States
- RC1DK086410/DK/NIDDK NIH HHS/United States
- R01 GM065386/GM/NIGMS NIH HHS/United States
- RC1 DK086410/DK/NIDDK NIH HHS/United States
- 5T32GM08349/GM/NIGMS NIH HHS/United States
- R37 GM059785/GM/NIGMS NIH HHS/United States
- AG038679/AG/NIA NIH HHS/United States
- R01 GM080148/GM/NIGMS NIH HHS/United States
- T32 GM008349/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

