pH-induced gene regulation of solvent production by Clostridium acetobutylicum in continuous culture: parameter estimation and sporulation modelling
- PMID: 23201580
- PMCID: PMC3547174
- DOI: 10.1016/j.mbs.2012.11.004
pH-induced gene regulation of solvent production by Clostridium acetobutylicum in continuous culture: parameter estimation and sporulation modelling
Abstract
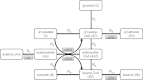
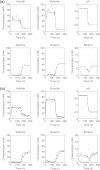
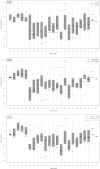
The acetone-butanol (AB) fermentation process in the anaerobic endospore-forming Gram-positive bacterium Clostridium acetobutylicum is useful as a producer of biofuels, particularly butanol. Recent work has concentrated on trying to improve the efficiency of the fermentation method, either through changes in the environmental conditions or by modifying the genome to selectively favour the production of one particular solvent over others. Fermentation of glucose by C. acetobutylicum occurs in two stages: initially the acids acetate and butyrate are produced and excreted and then, as the external pH falls, acetate and butyrate are ingested and further metabolised into the solvents acetone, butanol and ethanol. In order to optimise butanol production, it is important to understand how pH affects the enzyme-controlled reactions in the metabolism process. We adapt an ordinary differential equation model of the metabolic network with regulation at the genetic level for the required enzymes; parametrising the model using experimental data generated from continuous culture, we improve on previous point predictions (S. Haus, S. Jabbari, T. Millat, H. Janssen, R.-J. Fisher, H. Bahl, J. R. King, O. Wolkenhauer, A systems biology approach to investigate the effect of pH-induced gene regulation on solvent production by Clostridium acetobutylicum in continuous culture, BMC Systems Biology 5 (2011)) [1] both by using a different optimisation approach and by computing confidence intervals and correlation coefficients. We find in particular that the parameters are ill-determined from the data and that two separate clusters of parameters appear correlated, reflecting the importance of two metabolic intermediates. We extend the model further to include another aspect of the clostridial survival mechanism, sporulation, and by computation of the Akaike Information Criterion values find that the there is some evidence for the presence of sporulation during the shift.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures











References
-
- Gottwald M., Gottschalk G. The internal pH of Clostridium acetobutylicum and its effect on the shift from acid to solvent formation. Archives of Microbiology. 1985;143
-
- Dürre P., Fischer R.-J., Kuhn A., Lorenz K., Schreiber W., Stürzenhofecker B., Ullmann S., Winzer K., Sauer U. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties genetic organization and transcriptional regulation. FEMS Microbiology Reviews. 1995;17 - PubMed
-
- Bahl H., Andersch W., Braun K., Gottschalk G. Effect of pH and butyrate concentration on the production of acetone and butanol by Clostridium acetobutylicum grown in continuous culture. European Journal of Applied Microbiology and Biotechnology. 1982;14
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

