Control of somatic tissue differentiation by the long non-coding RNA TINCR
- PMID: 23201690
- PMCID: PMC3674581
- DOI: 10.1038/nature11661
Control of somatic tissue differentiation by the long non-coding RNA TINCR
Abstract
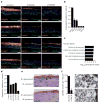
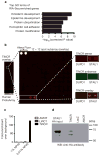
Several of the thousands of human long non-coding RNAs (lncRNAs) have been functionally characterized; however, potential roles for lncRNAs in somatic tissue differentiation remain poorly understood. Here we show that a 3.7-kilobase lncRNA, terminal differentiation-induced ncRNA (TINCR), controls human epidermal differentiation by a post-transcriptional mechanism. TINCR is required for high messenger RNA abundance of key differentiation genes, many of which are mutated in human skin diseases, including FLG, LOR, ALOXE3, ALOX12B, ABCA12, CASP14 and ELOVL3. TINCR-deficient epidermis lacked terminal differentiation ultrastructure, including keratohyalin granules and intact lamellar bodies. Genome-scale RNA interactome analysis revealed that TINCR interacts with a range of differentiation mRNAs. TINCR-mRNA interaction occurs through a 25-nucleotide 'TINCR box' motif that is strongly enriched in interacting mRNAs and required for TINCR binding. A high-throughput screen to analyse TINCR binding capacity to approximately 9,400 human recombinant proteins revealed direct binding of TINCR RNA to the staufen1 (STAU1) protein. STAU1-deficient tissue recapitulated the impaired differentiation seen with TINCR depletion. Loss of UPF1 and UPF2, both of which are required for STAU1-mediated RNA decay, however, did not have differentiation effects. Instead, the TINCR-STAU1 complex seems to mediate stabilization of differentiation mRNAs, such as KRT80. These data identify TINCR as a key lncRNA required for somatic tissue differentiation, which occurs through lncRNA binding to differentiation mRNAs to ensure their expression.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

