Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells
- PMID: 23202270
- PMCID: PMC3764492
- DOI: 10.1038/ni.2490
Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells
Abstract
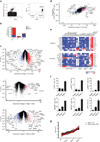
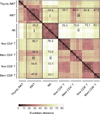
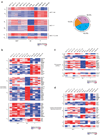
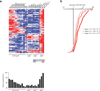
Invariant natural killer T cells (iNKT cells) are innate-like T lymphocytes that act as critical regulators of the immune response. To better characterize this population, we profiled gene expression in iNKT cells during ontogeny and in peripheral subsets as part of the Immunological Genome Project. High-resolution comparative transcriptional analyses defined developmental and subset-specific programs of gene expression by iNKT cells. In addition, we found that iNKT cells shared an extensive transcriptional program with NK cells, similar in magnitude to that shared with major histocompatibility complex (MHC)-restricted T cells. Notably, the program shared by NK cells and iNKT cells also operated constitutively in γδ T cells and in adaptive T cells after activation. Together our findings highlight a core effector program regulated distinctly in innate and adaptive lymphocytes.
Figures

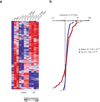
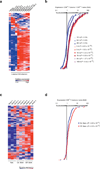
References
-
- Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. - PubMed
-
- Cohen NR, Garg S, Brenner MB. Antigen Presentation by CD1 Lipids, T Cells, and NKT Cells in Microbial Immunity. Adv Immunol. 2009;102:1–94. - PubMed
-
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. - PubMed
-
- Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11:197–206. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

