Short toxin-like proteins abound in Cnidaria genomes
- PMID: 23202321
- PMCID: PMC3509713
- DOI: 10.3390/toxins4111367
Short toxin-like proteins abound in Cnidaria genomes
Abstract
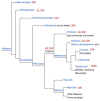
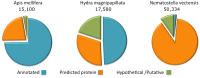
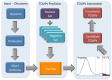
Cnidaria is a rich phylum that includes thousands of marine species. In this study, we focused on Anthozoa and Hydrozoa that are represented by the Nematostella vectensis (Sea anemone) and Hydra magnipapillata genomes. We present a method for ranking the toxin-like candidates from complete proteomes of Cnidaria. Toxin-like functions were revealed using ClanTox, a statistical machine-learning predictor trained on ion channel inhibitors from venomous animals. Fundamental features that were emphasized in training ClanTox include cysteines and their spacing along the sequences. Among the 83,000 proteins derived from Cnidaria representatives, we found 170 candidates that fulfill the properties of toxin-like-proteins, the vast majority of which were previously unrecognized as toxins. An additional 394 short proteins exhibit characteristics of toxin-like proteins at a moderate degree of confidence. Remarkably, only 11% of the predicted toxin-like proteins were previously classified as toxins. Based on our prediction methodology and manual annotation, we inferred functions for over 400 of these proteins. Such functions include protease inhibitors, membrane pore formation, ion channel blockers and metal binding proteins. Many of the proteins belong to small families of paralogs. We conclude that the evolutionary expansion of toxin-like proteins in Cnidaria contributes to their fitness in the complex environment of the aquatic ecosystem.
Figures

Similar articles
-
Expansion of tandem repeats in sea anemone Nematostella vectensis proteome: A source for gene novelty?BMC Genomics. 2009 Dec 10;10:593. doi: 10.1186/1471-2164-10-593. BMC Genomics. 2009. PMID: 20003297 Free PMC article.
-
ClanTox: a classifier of short animal toxins.Nucleic Acids Res. 2009 Jul;37(Web Server issue):W363-8. doi: 10.1093/nar/gkp299. Epub 2009 May 8. Nucleic Acids Res. 2009. PMID: 19429697 Free PMC article.
-
Identification of rDNA-specific non-LTR retrotransposons in Cnidaria.Mol Biol Evol. 2006 Oct;23(10):1984-93. doi: 10.1093/molbev/msl067. Epub 2006 Jul 26. Mol Biol Evol. 2006. PMID: 16870681
-
Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an overview.Mar Drugs. 2012 Aug;10(8):1812-1851. doi: 10.3390/md10081812. Epub 2012 Aug 22. Mar Drugs. 2012. PMID: 23015776 Free PMC article. Review.
-
Pore-forming toxins in Cnidaria.Semin Cell Dev Biol. 2017 Dec;72:133-141. doi: 10.1016/j.semcdb.2017.07.026. Epub 2017 Jul 24. Semin Cell Dev Biol. 2017. PMID: 28751252 Review.
Cited by
-
Overlooked Short Toxin-Like Proteins: A Shortcut to Drug Design.Toxins (Basel). 2017 Oct 29;9(11):350. doi: 10.3390/toxins9110350. Toxins (Basel). 2017. PMID: 29109389 Free PMC article.
-
An evolutionary timeline of the oxytocin signaling pathway.Commun Biol. 2024 Apr 17;7(1):471. doi: 10.1038/s42003-024-06094-9. Commun Biol. 2024. PMID: 38632466 Free PMC article.
References
-
- Bridge D., Cunningham C.W., DeSalle R., Buss L.W. Class-level relationships in the phylum Cnidaria: Molecular and morphological evidence. Mol. Biol. Evol. 1995;12:679–689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

