D471G mutation in LCMV-NP affects its ability to self-associate and results in a dominant negative effect in viral RNA synthesis
- PMID: 23202457
- PMCID: PMC3497045
- DOI: 10.3390/v4102137
D471G mutation in LCMV-NP affects its ability to self-associate and results in a dominant negative effect in viral RNA synthesis
Abstract
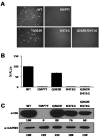
Arenaviruses merit significant interest because several family members are etiological agents of severe hemorrhagic fevers, representing a major burden to public health. Currently, there are no FDA-licensed vaccines against arenaviruses and the only available antiviral therapy is limited to the use of ribavirin that is partially effective. Arenavirus nucleoprotein (NP) is found associated with the genomic RNA forming the viral ribonucleoproteins (vRNPs) that together with the polymerase (L) direct viral replication and transcription. Virion formation requires the recruitment of vRNPs into budding sites, a process in which the arenavirus matrix-like protein (Z) plays a major role. Therefore, proper NP-NP and NP-Z interactions are required for the generation of infectious progeny. In this work we demonstrate the role of the amino acid residue D471 in the self-association of lymphocytic choriomeningitis virus nucleoprotein (LCMV-NP). Amino acid substitutions at this position abrogate NP oligomerization, affecting its ability to mediate replication and transcription of a minigenome reporter plasmid. However, its ability to interact with the Z protein, counteract the cellular interferon response and bind to dsRNA analogs was retained. Additionally, we also document the dominant negative effect of D471G mutation on viral infection, suggesting that NP self-association is an excellent target for the development of new antivirals against arenaviruses.
Figures





Similar articles
-
The C-terminal region of lymphocytic choriomeningitis virus nucleoprotein contains distinct and segregable functional domains involved in NP-Z interaction and counteraction of the type I interferon response.J Virol. 2011 Dec;85(24):13038-48. doi: 10.1128/JVI.05834-11. Epub 2011 Oct 5. J Virol. 2011. PMID: 21976642 Free PMC article.
-
Single nucleoprotein residue modulates arenavirus replication complex formation.mBio. 2015 Apr 28;6(3):e00524-15. doi: 10.1128/mBio.00524-15. mBio. 2015. PMID: 25922393 Free PMC article.
-
Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function.J Virol. 2012 Mar;86(6):3307-17. doi: 10.1128/JVI.05503-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258244 Free PMC article.
-
Multifunctional nature of the arenavirus RING finger protein Z.Viruses. 2012 Nov 9;4(11):2973-3011. doi: 10.3390/v4112973. Viruses. 2012. PMID: 23202512 Free PMC article. Review.
-
Molecular mechanism of arenavirus assembly and budding.Viruses. 2012 Oct 10;4(10):2049-79. doi: 10.3390/v4102049. Viruses. 2012. PMID: 23202453 Free PMC article. Review.
Cited by
-
Differential contributions of tacaribe arenavirus nucleoprotein N-terminal and C-terminal residues to nucleocapsid functional activity.J Virol. 2014 Jun;88(11):6492-505. doi: 10.1128/JVI.00321-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696466 Free PMC article.
-
Development of live-attenuated arenavirus vaccines based on codon deoptimization of the viral glycoprotein.Virology. 2017 Jan 15;501:35-46. doi: 10.1016/j.virol.2016.11.001. Epub 2016 Nov 14. Virology. 2017. PMID: 27855284 Free PMC article.
-
Reverse Genetics Approaches to Control Arenavirus.Methods Mol Biol. 2016;1403:313-51. doi: 10.1007/978-1-4939-3387-7_17. Methods Mol Biol. 2016. PMID: 27076139 Free PMC article.
-
Plasmid-Based Lassa Virus Reverse Genetics.Methods Mol Biol. 2024;2733:115-131. doi: 10.1007/978-1-0716-3533-9_8. Methods Mol Biol. 2024. PMID: 38064030
-
Arenavirus reverse genetics for vaccine development.J Gen Virol. 2013 Jun;94(Pt 6):1175-1188. doi: 10.1099/vir.0.051102-0. Epub 2013 Jan 30. J Gen Virol. 2013. PMID: 23364194 Free PMC article.
References
-
- Buchmeier M.J., Peters C.J., de la Torre J.C. Arenaviridae: The viruses and their replication. In: Fields B. N.; Knipe, D. M.; Howley P. M., editors. Fields virology. Fifth. Vol. 2. 2007. pp. 1792–1827.
-
- McCormick J.B., Fisher-Hoch S.P. Lassa Fever. In: Oldstone M. B., editor. Arenaviruses I. Vol. 262. Springer-Verlag; Berlin, Heidelberg, New York: 2002. pp. 75–110.
-
- Peters C.J. Human infection with Arenaviruses in the Americas. In: Oldstone M. B., editor. Arenaviruses I. Vol. 262. Springer-Verlag; Berlin, Heidelberg, New York: 2002. pp. 65–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

