Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems
- PMID: 23202464
- PMCID: PMC3497052
- DOI: 10.3390/v4102291
Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems
Abstract
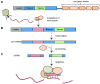
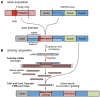
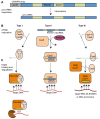
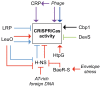
Phages are the most abundant biological entities on earth and pose a constant challenge to their bacterial hosts. Thus, bacteria have evolved numerous 'innate' mechanisms of defense against phage, such as abortive infection or restriction/modification systems. In contrast, the clustered regularly interspaced short palindromic repeats (CRISPR) systems provide acquired, yet heritable, sequence-specific 'adaptive' immunity against phage and other horizontally-acquired elements, such as plasmids. Resistance is acquired following viral infection or plasmid uptake when a short sequence of the foreign genome is added to the CRISPR array. CRISPRs are then transcribed and processed, generally by CRISPR associated (Cas) proteins, into short interfering RNAs (crRNAs), which form part of a ribonucleoprotein complex. This complex guides the crRNA to the complementary invading nucleic acid and targets this for degradation. Recently, there have been rapid advances in our understanding of CRISPR/Cas systems. In this review, we will present the current model(s) of the molecular events involved in both the acquisition of immunity and interference stages and will also address recent progress in our knowledge of the regulation of CRISPR/Cas systems.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

