Modelling translation initiation under the influence of sRNA
- PMID: 23203192
- PMCID: PMC3546686
- DOI: 10.3390/ijms131216223
Modelling translation initiation under the influence of sRNA
Abstract
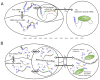
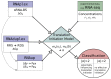
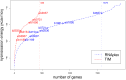
Bacterial small non-coding RNA (sRNA) plays an important role in post-transcriptional gene regulation. Although the number of annotated sRNA is steadily increasing, their functional characterization is still lagging behind. Various computational strategies for finding sRNA−mRNA interactions, and thus putative sRNA targets, were developed. Most of them suffer from a high false positive rate. Here, we present a qualitative model to simulate the effect of an sRNA on the translation initiation of a potential target. Information about the ribosome−mRNA interaction, sRNA−mRNA interaction and expression information from deep sequencing experiments is integrated to calculate the change in translation initiation complex formation, as a proxy for translational activity. This model can be used to post-evaluate predicted targets, hence condensing the list of potential targets. We show that our translation initiation model, under the influence of an sRNA, can successfully simulate thirteen out of fifteen tested sRNA−mRNA interactions in a qualitative manner. To show the gain in specificity, we applied our method to a target search for the Escherichia coli sRNA RyhB. Compared with simple target prediction without post-evaluation, we reduce the number of targets to less than one fourth potential targets, considerably reducing the burden of experimental validation.
Figures
Similar articles
-
Identification of bacterial sRNA regulatory targets using ribosome profiling.Nucleic Acids Res. 2015 Dec 2;43(21):10308-20. doi: 10.1093/nar/gkv1158. Epub 2015 Nov 5. Nucleic Acids Res. 2015. PMID: 26546513 Free PMC article.
-
sRNA Target Prediction Organizing Tool (SPOT) Integrates Computational and Experimental Data To Facilitate Functional Characterization of Bacterial Small RNAs.mSphere. 2019 Jan 30;4(1):e00561-18. doi: 10.1128/mSphere.00561-18. mSphere. 2019. PMID: 30700509 Free PMC article.
-
The sRNA RyhB regulates the synthesis of the Escherichia coli methionine sulfoxide reductase MsrB but not MsrA.PLoS One. 2013 May 9;8(5):e63647. doi: 10.1371/journal.pone.0063647. Print 2013. PLoS One. 2013. PMID: 23671689 Free PMC article.
-
New insights into small RNA-dependent translational regulation in prokaryotes.Trends Genet. 2013 Feb;29(2):92-8. doi: 10.1016/j.tig.2012.10.004. Epub 2012 Nov 6. Trends Genet. 2013. PMID: 23141721 Review.
-
Bacterial small RNA-based negative regulation: Hfq and its accomplices.J Biol Chem. 2013 Mar 22;288(12):7996-8003. doi: 10.1074/jbc.R112.441386. Epub 2013 Jan 29. J Biol Chem. 2013. PMID: 23362267 Free PMC article. Review.
Cited by
-
Small RNA-mediated Cry toxin silencing allows Bacillus thuringiensis to evade Caenorhabditis elegans avoidance behavioral defenses.Nucleic Acids Res. 2018 Jan 9;46(1):159-173. doi: 10.1093/nar/gkx959. Nucleic Acids Res. 2018. PMID: 29069426 Free PMC article.
-
Sequence Design for RNA-RNA Interactions.Methods Mol Biol. 2025;2847:1-16. doi: 10.1007/978-1-0716-4079-1_1. Methods Mol Biol. 2025. PMID: 39312133
-
Hybrid semiparametric systems for quantitative sequence-activity modeling of synthetic biological parts.Synth Biol (Oxf). 2018 Jun 26;3(1):ysy010. doi: 10.1093/synbio/ysy010. eCollection 2018. Synth Biol (Oxf). 2018. PMID: 32995518 Free PMC article.
-
A Canonical Biophysical Model of the CsrA Global Regulator Suggests Flexible Regulator-Target Interactions.Sci Rep. 2018 Jul 2;8(1):9892. doi: 10.1038/s41598-018-27474-2. Sci Rep. 2018. PMID: 29967470 Free PMC article.
-
In silico estimation of translation efficiency in human cell lines: potential evidence for widespread translational control.PLoS One. 2013;8(2):e57625. doi: 10.1371/journal.pone.0057625. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23460887 Free PMC article.
References
-
- Backofen R., Hess W. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

