RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more
- PMID: 23203884
- PMCID: PMC3531196
- DOI: 10.1093/nar/gks1201
RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more
Abstract
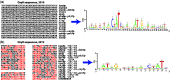
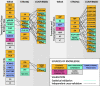
This article summarizes our progress with RegulonDB (http://regulondb.ccg.unam.mx/) during the past 2 years. We have kept up-to-date the knowledge from the published literature regarding transcriptional regulation in Escherichia coli K-12. We have maintained and expanded our curation efforts to improve the breadth and quality of the encoded experimental knowledge, and we have implemented criteria for the quality of our computational predictions. Regulatory phrases now provide high-level descriptions of regulatory regions. We expanded the assignment of quality to various sources of evidence, particularly for knowledge generated through high-throughput (HT) technology. Based on our analysis of most relevant methods, we defined rules for determining the quality of evidence when multiple independent sources support an entry. With this latest release of RegulonDB, we present a new highly reliable larger collection of transcription start sites, a result of our experimental HT genome-wide efforts. These improvements, together with several novel enhancements (the tracks display, uploading format and curational guidelines), address the challenges of incorporating HT-generated knowledge into RegulonDB. Information on the evolutionary conservation of regulatory elements is also available now. Altogether, RegulonDB version 8.0 is a much better home for integrating knowledge on gene regulation from the sources of information currently available.
Figures



References
-
- Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. - PMC - PubMed
-
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 2001;305:673–688. - PubMed
-
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 2001;305:689–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

