Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing
- PMID: 23204167
- PMCID: PMC3549603
- DOI: 10.1093/infdis/jis734
Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing
Abstract
Background: Clostridium difficile is a major cause of nosocomial diarrhea, with 30-day mortality reaching 30%. The cell surface comprises a paracrystalline proteinaceous S-layer encoded by the slpA gene within the cell wall protein (cwp) gene cluster. Our purpose was to understand the diversity and evolution of slpA and nearby genes also encoding immunodominant cell surface antigens.
Methods: Whole-genome sequences were determined for 57 C. difficile isolates representative of the population structure and different clinical phenotypes. Phylogenetic analyses were performed on their genomic region (>63 kb) spanning the cwp cluster.
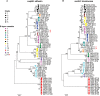
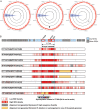
Results: Genetic diversity across the cwp cluster peaked within slpA, cwp66 (adhesin), and secA2 (secretory translocase). These genes formed a 10-kb cassette, of which 12 divergent variants were found. Homologous recombination involving this cassette caused it to associate randomly with genotype. One cassette contained a novel insertion (length, approximately 24 kb) that resembled S-layer glycosylation gene clusters.
Conclusions: Genetic exchange of S-layer cassettes parallels polysaccharide capsular switching in other species. Both cause major antigenic shifts, while the remainder of the genome is unchanged. C. difficile genotype is therefore not predictive of antigenic type. S-layer switching and immune escape could help explain temporal and geographic variation in C. difficile epidemiology and may inform genotyping and vaccination strategies.
Figures


References
-
- Mayer LW, Reeves MW, Al-Hamdan N, et al. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J Infect Dis. 2002;185:1596–605. - PubMed
-
- Pallen MJ, Loman NJ, Penn CW. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr Opin Microbiol. 2010;13:625–31. - PubMed
-
- Bauer MP, Notermans DW, van Benthem BH, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

