A novel in silico approach to identify potential therapeutic targets in human bacterial pathogens
- PMID: 23205162
- PMCID: PMC3238024
- DOI: 10.1007/s11568-011-9152-7
A novel in silico approach to identify potential therapeutic targets in human bacterial pathogens
Abstract
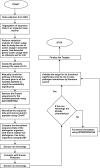
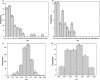
In recent years, genome-sequencing projects of pathogens and humans have revolutionized microbial drug target identification. Of the several known genomic strategies, subtractive genomics has been successfully utilized for identifying microbial drug targets. The present work demonstrates a novel genomics approach in which codon adaptation index (CAI), a measure used to predict the translational efficiency of a gene based on synonymous codon usage, is coupled with subtractive genomics approach for mining potential drug targets. The strategy adopted is demonstrated using respiratory pathogens, namely, Streptococcus pneumoniae and Haemophilus influenzae as examples. Our approach identified 8 potent target genes (Streptococcus pneumoniae-2, H. influenzae-6), which are functionally significant and also play key role in host-pathogen interactions. This approach facilitates swift identification of potential drug targets, thereby enabling the search for new inhibitors. These results underscore the utility of CAI for enhanced in silico drug target identification.
Electronic supplementary material: The online version of this article (doi:10.1007/s11568-011-9152-7) contains supplementary material, which is available to authorized users.
Keywords: Bacterial pathogens; CAI value; Codon usage; Drug targets; Subtractive genomics.
Figures
Similar articles
-
Identification of potential therapeutic targets in Neisseria gonorrhoeae by an in-silico approach.J Theor Biol. 2020 Apr 7;490:110172. doi: 10.1016/j.jtbi.2020.110172. Epub 2020 Jan 20. J Theor Biol. 2020. PMID: 31972174
-
Biocomputational strategies for microbial drug target identification.Methods Mol Med. 2008;142:1-9. doi: 10.1007/978-1-59745-246-5_1. Methods Mol Med. 2008. PMID: 18437301
-
Target identification in Fusobacterium nucleatum by subtractive genomics approach and enrichment analysis of host-pathogen protein-protein interactions.BMC Microbiol. 2016 May 12;16:84. doi: 10.1186/s12866-016-0700-0. BMC Microbiol. 2016. PMID: 27176600 Free PMC article.
-
Subtractive genomics approach: A guide to unveiling therapeutic targets across pathogens.J Microbiol Methods. 2025 Jul;232-234:107127. doi: 10.1016/j.mimet.2025.107127. Epub 2025 Apr 7. J Microbiol Methods. 2025. PMID: 40204082 Review.
-
Microbial interactions in the respiratory tract.Pediatr Infect Dis J. 2009 Oct;28(10 Suppl):S121-6. doi: 10.1097/INF.0b013e3181b6d7ec. Pediatr Infect Dis J. 2009. PMID: 19918134 Review.
Cited by
-
Drug repurposing for the treatment of staphylococcal infections.Curr Pharm Des. 2015;21(16):2089-100. doi: 10.2174/1381612821666150310104416. Curr Pharm Des. 2015. PMID: 25760334 Free PMC article. Review.
-
Pathogenomic in silico approach identifies NSP-A and Fe-IIISBP as possible drug targets in Neisseria Meningitidis MC58 and development of pharmacophores as novel therapeutic candidates.Mol Divers. 2023 Jun;27(3):1163-1184. doi: 10.1007/s11030-022-10480-y. Epub 2022 Jul 25. Mol Divers. 2023. PMID: 35879631
-
Finding Potential Therapeutic Targets against Shigella flexneri through Proteome Exploration.Front Microbiol. 2016 Nov 22;7:1817. doi: 10.3389/fmicb.2016.01817. eCollection 2016. Front Microbiol. 2016. PMID: 27920755 Free PMC article.
-
In silico characterization of hypothetical proteins from Orientia tsutsugamushi str. Karp uncovers virulence genes.Heliyon. 2019 Nov 1;5(10):e02734. doi: 10.1016/j.heliyon.2019.e02734. eCollection 2019 Oct. Heliyon. 2019. PMID: 31720472 Free PMC article.
-
Genome scale identification, structural analysis, and classification of periplasmic binding proteins from Mycobacterium tuberculosis.Curr Genet. 2017 Jun;63(3):553-576. doi: 10.1007/s00294-016-0664-5. Epub 2016 Nov 17. Curr Genet. 2017. PMID: 27858159
References
LinkOut - more resources
Full Text Sources

