Cernunnos deficiency reduces thymocyte life span and alters the T cell repertoire in mice and humans
- PMID: 23207905
- PMCID: PMC3571340
- DOI: 10.1128/MCB.01057-12
Cernunnos deficiency reduces thymocyte life span and alters the T cell repertoire in mice and humans
Abstract
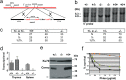
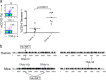
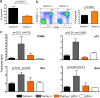
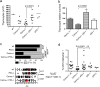
Cernunnos is a DNA repair factor of the nonhomologous end-joining machinery. Its deficiency in humans causes radiosensitive severe combined immune deficiency (SCID) with microcephaly, characterized in part by a profound lymphopenia. In contrast to the human condition, the immune system of Cernunnos knockout (KO) mice is not overwhelmingly affected. In particular, Cernunnos is dispensable during V(D)J recombination in lymphoid cells. Nevertheless, the viability of thymocytes is reduced in Cernunnos KO mice, owing to the chronic activation of a P53-dependent DNA damage response. This translates into a qualitative alteration of the T cell repertoire to one in which the most distal Vα and Jα segments are missing. This results in the contraction of discrete T cell populations, such as invariant natural killer T (iNKT) and mucosa-associated invariant T (MAIT) cells, in both humans and mice.
Figures





References
-
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124:287–299 - PubMed
-
- Ahnesorg P, Smith P, Jackson SP. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124:301–313 - PubMed
-
- Andres SN, Modesti M, Tsai CJ, Chu G, Junop MS. 2007. Crystal structure of human XLF: a twist in nonhomologous DNA end-joining. Mol. Cell 28:1093–1101 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
