Rho and NusG suppress pervasive antisense transcription in Escherichia coli
- PMID: 23207917
- PMCID: PMC3521622
- DOI: 10.1101/gad.196741.112
Rho and NusG suppress pervasive antisense transcription in Escherichia coli
Abstract
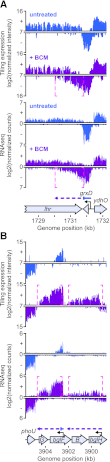
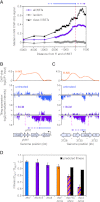
Despite the prevalence of antisense transcripts in bacterial transcriptomes, little is known about how their synthesis is controlled. We report that a major function of the Escherichia coli termination factor Rho and its cofactor, NusG, is suppression of ubiquitous antisense transcription genome-wide. Rho binds C-rich unstructured nascent RNA (high C/G ratio) prior to its ATP-dependent dissociation of transcription complexes. NusG is required for efficient termination at minority subsets (~20%) of both antisense and sense Rho-dependent terminators with lower C/G ratio sequences. In contrast, a widely studied nusA deletion proposed to compromise Rho-dependent termination had no effect on antisense or sense Rho-dependent terminators in vivo. Global colocalization of the histone-like nucleoid-structuring protein (H-NS) with Rho-dependent terminators and genetic interactions between hns and rho suggest that H-NS aids Rho in suppression of antisense transcription. The combined actions of Rho, NusG, and H-NS appear to be analogous to the Sen1-Nrd1-Nab3 and nucleosome systems that suppress antisense transcription in eukaryotes.
Figures





Comment in
-
Bacterial physiology: Antisense suppression.Nat Rev Microbiol. 2013 Feb;11(2):72. doi: 10.1038/nrmicro2961. Nat Rev Microbiol. 2013. PMID: 23321527 No abstract available.
References
-
- Adhya S, Gottesman M, De Crombrugghe B 1974. Termination and antitermination in transcription: Control of gene expression. Basic Life Sci 3: 213–221 - PubMed
-
- Arigo JT, Eyler DE, Carroll KL, Corden JL 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 - PubMed
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
