A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington's disease
- PMID: 23209424
- PMCID: PMC3510027
- DOI: 10.1371/journal.pgen.1003042
A genome-scale RNA-interference screen identifies RRAS signaling as a pathologic feature of Huntington's disease
Abstract
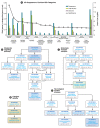
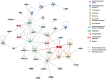
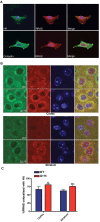
A genome-scale RNAi screen was performed in a mammalian cell-based assay to identify modifiers of mutant huntingtin toxicity. Ontology analysis of suppressor data identified processes previously implicated in Huntington's disease, including proteolysis, glutamate excitotoxicity, and mitochondrial dysfunction. In addition to established mechanisms, the screen identified multiple components of the RRAS signaling pathway as loss-of-function suppressors of mutant huntingtin toxicity in human and mouse cell models. Loss-of-function in orthologous RRAS pathway members also suppressed motor dysfunction in a Drosophila model of Huntington's disease. Abnormal activation of RRAS and a down-stream effector, RAF1, was observed in cellular models and a mouse model of Huntington's disease. We also observe co-localization of RRAS and mutant huntingtin in cells and in mouse striatum, suggesting that activation of R-Ras may occur through protein interaction. These data indicate that mutant huntingtin exerts a pathogenic effect on this pathway that can be corrected at multiple intervention points including RRAS, FNTA/B, PIN1, and PLK1. Consistent with these results, chemical inhibition of farnesyltransferase can also suppress mutant huntingtin toxicity. These data suggest that pharmacological inhibition of RRAS signaling may confer therapeutic benefit in Huntington's disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- The Huntington's Disease Collaborative Research Group, Cell 72, 971 (Mar 26, 1993). - PubMed
-
- de la Monte SM, Vonsattel JP, Richardson EP Jr (1988) Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington's disease. J Neuropathol Exp Neurol 47: 516–525. - PubMed
-
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, et al. (2001) Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293: 493–498. - PubMed
-
- Cha CI, Chung YH, Shin CM, Shin DH, Kim YS, et al. (2000) Immunocytochemical study on the distribution of nitrotyrosine in the brain of the transgenic mice expressing a human Cu/Zn SOD mutation. Brain Res 853: 156–161. - PubMed
-
- Weydt P, Pineda VV, Torrence AE, Libby RT, Satterfield TF, et al. (2006) Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab 4: 349–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG000266/AG/NIA NIH HHS/United States
- NIH R01 LM009722/LM/NLM NIH HHS/United States
- T32 AG000266/AG/NIA NIH HHS/United States
- U54 HG004028/HG/NHGRI NIH HHS/United States
- NIH NS055247/NS/NINDS NIH HHS/United States
- NIH NS042179/NS/NINDS NIH HHS/United States
- T32 NS043124/NS/NINDS NIH HHS/United States
- R01 NS042179/NS/NINDS NIH HHS/United States
- R56 NS042179/NS/NINDS NIH HHS/United States
- P30 AG025708/AG/NIA NIH HHS/United States
- R01 NS055247/NS/NINDS NIH HHS/United States
- NIH U54-HG004028/HG/NHGRI NIH HHS/United States
- R01 LM009722/LM/NLM NIH HHS/United States
- NIH NS040251/NS/NINDS NIH HHS/United States
- P30AG025708/AG/NIA NIH HHS/United States
- R01 NS040251/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

