Tissue-specific regulation of chromatin insulator function
- PMID: 23209434
- PMCID: PMC3510032
- DOI: 10.1371/journal.pgen.1003069
Tissue-specific regulation of chromatin insulator function
Abstract
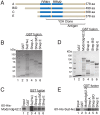
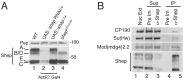
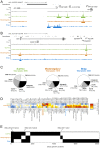
Chromatin insulators organize the genome into distinct transcriptional domains and contribute to cell type-specific chromatin organization. However, factors regulating tissue-specific insulator function have not yet been discovered. Here we identify the RNA recognition motif-containing protein Shep as a direct interactor of two individual components of the gypsy insulator complex in Drosophila. Mutation of shep improves gypsy-dependent enhancer blocking, indicating a role as a negative regulator of insulator activity. Unlike ubiquitously expressed core gypsy insulator proteins, Shep is highly expressed in the central nervous system (CNS) with lower expression in other tissues. We developed a novel, quantitative tissue-specific barrier assay to demonstrate that Shep functions as a negative regulator of insulator activity in the CNS but not in muscle tissue. Additionally, mutation of shep alters insulator complex nuclear localization in the CNS but has no effect in other tissues. Consistent with negative regulatory activity, ChIP-seq analysis of Shep in a CNS-derived cell line indicates substantial genome-wide colocalization with a single gypsy insulator component but limited overlap with intact insulator complexes. Taken together, these data reveal a novel, tissue-specific mode of regulation of a chromatin insulator.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713. - PubMed
-
- Pai CY, Lei EP, Ghosh D, Corces VG (2004) The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell 16: 737–748. - PubMed
-
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

